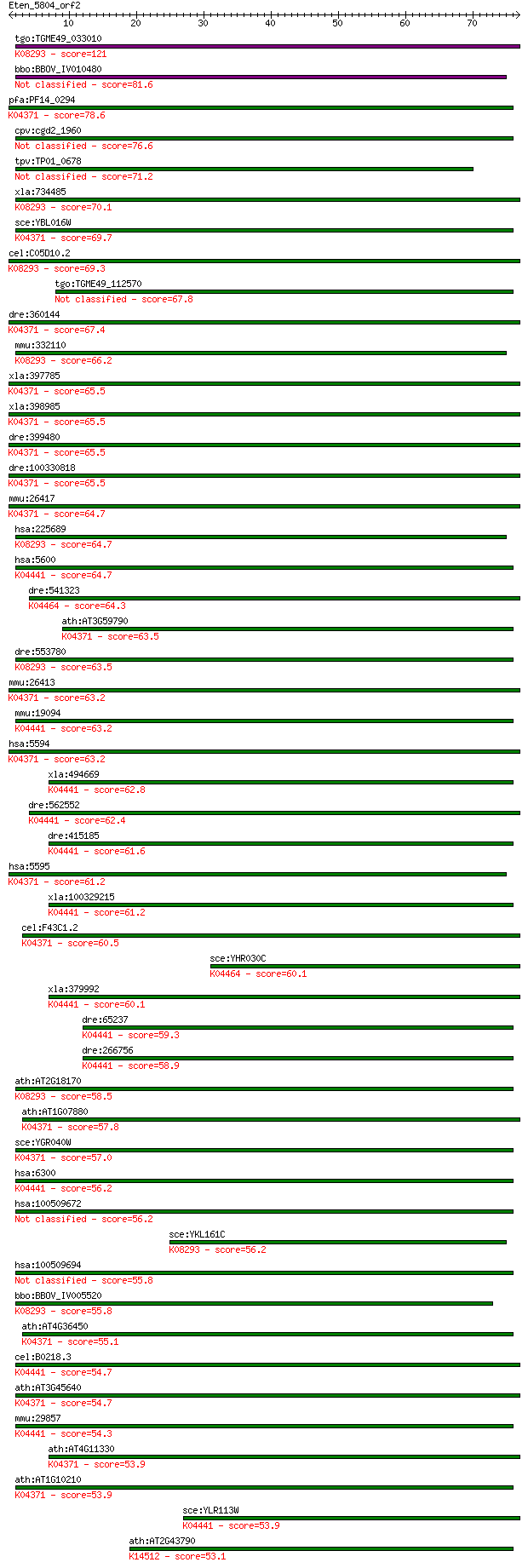
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5804_orf2
Length=76
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_033010 mitogen-activated protein kinase 2 (EC:2.7.1... 121 6e-28
bbo:BBOV_IV010480 23.m06413; mitogen activated protein kinase ... 81.6 6e-16
pfa:PF14_0294 PfMAP1; mitogen-activated protein kinase 1; K043... 78.6 5e-15
cpv:cgd2_1960 mitogen-activated protein kinase 1, serine/threo... 76.6 2e-14
tpv:TP01_0678 serine/threonine protein kinase 71.2 8e-13
xla:734485 mapk15, MGC99048; mitogen-activated protein kinase ... 70.1 2e-12
sce:YBL016W FUS3, DAC2; Fus3p (EC:2.7.11.24); K04371 extracell... 69.7 2e-12
cel:C05D10.2 hypothetical protein; K08293 mitogen-activated pr... 69.3 3e-12
tgo:TGME49_112570 mitogen-activated protein kinase, putative (... 67.8 9e-12
dre:360144 mapk1, zERK2; mitogen-activated protein kinase 1 (E... 67.4 1e-11
mmu:332110 Mapk15, BC048082, MGC56865, MGC56903; mitogen-activ... 66.2 2e-11
xla:397785 mapk1-b, Xp42, erk, erk2, mapk, mapk1b, mpk1; mitog... 65.5 4e-11
xla:398985 mapk1-a, MGC68934, erk, erk2, mapk, mapk1, mapk1a, ... 65.5 4e-11
dre:399480 mapk3, ERK1, fi06b09, wu:fi06b09, zERK1; mitogen-ac... 65.5 5e-11
dre:100330818 mitogen-activated protein kinase 3-like; K04371 ... 65.5 5e-11
mmu:26417 Mapk3, Erk-1, Erk1, Ert2, Esrk1, Mnk1, Mtap2k, Prkm3... 64.7 6e-11
hsa:225689 MAPK15, ERK7, ERK8; mitogen-activated protein kinas... 64.7 7e-11
hsa:5600 MAPK11, P38B, P38BETA2, PRKM11, SAPK2, SAPK2B, p38-2,... 64.7 8e-11
dre:541323 mapk7, bmk1, erk5, wu:fb73b02, zgc:113111; mitogen-... 64.3 9e-11
ath:AT3G59790 ATMPK10; MAP kinase/ kinase; K04371 extracellula... 63.5 1e-10
dre:553780 mapk15, MGC110345, erk7, zgc:110345; mitogen-activa... 63.5 2e-10
mmu:26413 Mapk1, 9030612K14Rik, AA407128, AU018647, C78273, ER... 63.2 2e-10
mmu:19094 Mapk11, DKFZp586C1322, P38b, Prkm11, Sapk2, Sapk2b, ... 63.2 2e-10
hsa:5594 MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2,... 63.2 2e-10
xla:494669 mapk11, p38-2, p38Beta, p38b, p38beta2, prkm11, sap... 62.8 2e-10
dre:562552 fc89f08, wu:fc89f08; zgc:171775; K04441 p38 MAP kin... 62.4 3e-10
dre:415185 zgc:86905; K04441 p38 MAP kinase [EC:2.7.11.24] 61.6 7e-10
hsa:5595 MAPK3, ERK1, HS44KDAP, HUMKER1A, MGC20180, P44ERK1, P... 61.2 7e-10
xla:100329215 p38-beta; mitogen-activated protein kinase p38 b... 61.2 8e-10
cel:F43C1.2 mpk-1; MAP Kinase family member (mpk-1); K04371 ex... 60.5 1e-09
sce:YHR030C SLT2, BYC2, MPK1, SLK2; Serine/threonine MAP kinas... 60.1 2e-09
xla:379992 mapk14, MGC69036, csbp, mapk14a, mxi2, p38, p38-alp... 60.1 2e-09
dre:65237 mapk14a, MAPK14, fk28c03, hm:zeh1243, p38, p38a, wu:... 59.3 3e-09
dre:266756 mapk14b, MAPK14, p38b, zp38b; mitogen-activated pro... 58.9 3e-09
ath:AT2G18170 ATMPK7; ATMPK7 (ARABIDOPSIS THALIANA MAP KINASE ... 58.5 5e-09
ath:AT1G07880 ATMPK13; MAP kinase/ kinase; K04371 extracellula... 57.8 7e-09
sce:YGR040W KSS1; Kss1p (EC:2.7.11.24); K04371 extracellular s... 57.0 2e-08
hsa:6300 MAPK12, ERK3, ERK6, P38GAMMA, PRKM12, SAPK-3, SAPK3; ... 56.2 2e-08
hsa:100509672 mitogen-activated protein kinase 12-like 56.2 2e-08
sce:YKL161C MLP1; Protein kinase implicated in the Slt2p mitog... 56.2 3e-08
hsa:100509694 mitogen-activated protein kinase 12-like 55.8 3e-08
bbo:BBOV_IV005520 23.m06268; mitogen-activated protein kinase;... 55.8 3e-08
ath:AT4G36450 ATMPK14 (Mitogen-activated protein kinase 14); M... 55.1 5e-08
cel:B0218.3 pmk-1; P38 Map Kinase family member (pmk-1); K0444... 54.7 7e-08
ath:AT3G45640 ATMPK3; ATMPK3 (ARABIDOPSIS THALIANA MITOGEN-ACT... 54.7 8e-08
mmu:29857 Mapk12, AW123708, Erk6, P38gamma, Prkm12, Sapk3; mit... 54.3 1e-07
ath:AT4G11330 ATMPK5; ATMPK5 (MAP KINASE 5); MAP kinase/ kinas... 53.9 1e-07
ath:AT1G10210 ATMPK1; ATMPK1 (MITOGEN-ACTIVATED PROTEIN KINASE... 53.9 1e-07
sce:YLR113W HOG1, SSK3; Hog1p (EC:2.7.11.24); K04441 p38 MAP k... 53.9 1e-07
ath:AT2G43790 ATMPK6; ATMPK6 (ARABIDOPSIS THALIANA MAP KINASE ... 53.1 2e-07
> tgo:TGME49_033010 mitogen-activated protein kinase 2 (EC:2.7.11.24);
K08293 mitogen-activated protein kinase [EC:2.7.11.24]
Length=669
Score = 121 bits (303), Expect = 6e-28, Method: Compositional matrix adjust.
Identities = 57/75 (76%), Positives = 64/75 (85%), Gaps = 0/75 (0%)
Query 2 EDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALE 61
EDV A++SPFAATMME LP+ K K +D FP+A P ALDLL+QLLQFNP KRISAEK LE
Sbjct 243 EDVDAVKSPFAATMMESLPLGKVKNFKDAFPNASPEALDLLKQLLQFNPNKRISAEKGLE 302
Query 62 HPYVRQFHNPEDEPV 76
HPYVRQFH+PEDEPV
Sbjct 303 HPYVRQFHSPEDEPV 317
> bbo:BBOV_IV010480 23.m06413; mitogen activated protein kinase
(EC:2.7.1.-)
Length=506
Score = 81.6 bits (200), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 37/73 (50%), Positives = 50/73 (68%), Gaps = 0/73 (0%)
Query 2 EDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALE 61
ED+ ++ SPF MM L V+ K +++YFP+AP A+DL+ +LLQFNP KRI+ +AL
Sbjct 239 EDIESLGSPFTTLMMGSLASVQNKPVKEYFPNAPDDAVDLVVRLLQFNPKKRITTLEALN 298
Query 62 HPYVRQFHNPEDE 74
HPYVR FH D
Sbjct 299 HPYVRSFHKSNDS 311
> pfa:PF14_0294 PfMAP1; mitogen-activated protein kinase 1; K04371
extracellular signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=914
Score = 78.6 bits (192), Expect = 5e-15, Method: Composition-based stats.
Identities = 38/75 (50%), Positives = 51/75 (68%), Gaps = 0/75 (0%)
Query 1 REDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKAL 60
++D+ IRSPFA ++ +K+K L+D A +LDLL +LLQFNP KRISAE AL
Sbjct 248 KKDIEDIRSPFAEKIISSFVDLKKKNLKDICYKASNESLDLLEKLLQFNPSKRISAENAL 307
Query 61 EHPYVRQFHNPEDEP 75
+H YV +FH+ DEP
Sbjct 308 KHKYVEEFHSIIDEP 322
> cpv:cgd2_1960 mitogen-activated protein kinase 1, serine/threonine
protein kinase
Length=710
Score = 76.6 bits (187), Expect = 2e-14, Method: Composition-based stats.
Identities = 43/90 (47%), Positives = 52/90 (57%), Gaps = 16/90 (17%)
Query 2 EDVAAIRSPFAATMMEGLPV---VKQKKLEDYF-------------PDAPPMALDLLRQL 45
EDV +I+SPFA TM+E L ++Q D F D ALDLL +L
Sbjct 255 EDVESIQSPFAKTMIESLKEKVEIRQSNKRDIFTKWKNLLLKINPKADCNEEALDLLDKL 314
Query 46 LQFNPLKRISAEKALEHPYVRQFHNPEDEP 75
LQFNP KRISA AL+HP+V FHNP +EP
Sbjct 315 LQFNPNKRISANDALKHPFVSIFHNPNEEP 344
> tpv:TP01_0678 serine/threonine protein kinase
Length=677
Score = 71.2 bits (173), Expect = 8e-13, Method: Composition-based stats.
Identities = 31/68 (45%), Positives = 45/68 (66%), Gaps = 1/68 (1%)
Query 2 EDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALE 61
ED+ ++ SPF M+ L +++K +++YFP+ LDLL +LLQFNP KRI+ AL
Sbjct 250 EDMDSLSSPFTKVMISSL-TIRKKNIKEYFPNTCEEGLDLLNRLLQFNPTKRINTVDALA 308
Query 62 HPYVRQFH 69
HPY+ FH
Sbjct 309 HPYLSNFH 316
> xla:734485 mapk15, MGC99048; mitogen-activated protein kinase
15 (EC:2.7.11.24); K08293 mitogen-activated protein kinase
[EC:2.7.11.24]
Length=586
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 34/76 (44%), Positives = 52/76 (68%), Gaps = 1/76 (1%)
Query 2 EDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPM-ALDLLRQLLQFNPLKRISAEKAL 60
ED+ +I+S + A+++ + + + + FP + P ALDLL +LL FNP KR++AE+AL
Sbjct 241 EDIVSIKSEYGASVISRMSSKHKVPMAELFPASCPREALDLLSKLLVFNPGKRLTAEEAL 300
Query 61 EHPYVRQFHNPEDEPV 76
EHPYV +FH+P EP
Sbjct 301 EHPYVSRFHSPAREPA 316
> sce:YBL016W FUS3, DAC2; Fus3p (EC:2.7.11.24); K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=353
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 33/74 (44%), Positives = 49/74 (66%), Gaps = 0/74 (0%)
Query 2 EDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALE 61
D+ I SP A ++ LP+ LE FP P +DLL+++L F+P KRI+A++ALE
Sbjct 246 NDLRCIESPRAREYIKSLPMYPAAPLEKMFPRVNPKGIDLLQRMLVFDPAKRITAKEALE 305
Query 62 HPYVRQFHNPEDEP 75
HPY++ +H+P DEP
Sbjct 306 HPYLQTYHDPNDEP 319
> cel:C05D10.2 hypothetical protein; K08293 mitogen-activated
protein kinase [EC:2.7.11.24]
Length=470
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 33/76 (43%), Positives = 52/76 (68%), Gaps = 0/76 (0%)
Query 1 REDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKAL 60
R D+A+I S +AA+++E +P +K L+ + A+D++++LL F P KR++ E+ L
Sbjct 242 RADIASIGSHYAASVLEKMPQRPRKPLDLIITQSQTAAIDMVQRLLIFAPQKRLTVEQCL 301
Query 61 EHPYVRQFHNPEDEPV 76
HPYV QFHNP +EPV
Sbjct 302 VHPYVVQFHNPSEEPV 317
> tgo:TGME49_112570 mitogen-activated protein kinase, putative
(EC:2.7.11.24)
Length=1212
Score = 67.8 bits (164), Expect = 9e-12, Method: Composition-based stats.
Identities = 35/68 (51%), Positives = 41/68 (60%), Gaps = 0/68 (0%)
Query 8 RSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALEHPYVRQ 67
RS A +E LP KLED FPDA ALDLL LL F+P KRI+ ++AL H Y
Sbjct 361 RSEAARRFIESLPNSDPYKLEDLFPDASKAALDLLSNLLTFDPAKRITVQEALRHEYFEG 420
Query 68 FHNPEDEP 75
H+ EDEP
Sbjct 421 LHSVEDEP 428
> dre:360144 mapk1, zERK2; mitogen-activated protein kinase 1
(EC:2.7.11.24); K04371 extracellular signal-regulated kinase
1/2 [EC:2.7.11.24]
Length=369
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 34/76 (44%), Positives = 48/76 (63%), Gaps = 0/76 (0%)
Query 1 REDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKAL 60
+ED+ I + A + LP+ + FP+A P ALDLL ++L FNP KRI E+AL
Sbjct 258 QEDLNCIINIKARNYLLSLPLRSKVPWNRLFPNADPKALDLLDKMLTFNPHKRIEVEEAL 317
Query 61 EHPYVRQFHNPEDEPV 76
HPY+ Q+++P DEPV
Sbjct 318 AHPYLEQYYDPTDEPV 333
> mmu:332110 Mapk15, BC048082, MGC56865, MGC56903; mitogen-activated
protein kinase 15 (EC:2.7.11.24); K08293 mitogen-activated
protein kinase [EC:2.7.11.24]
Length=549
Score = 66.2 bits (160), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 34/74 (45%), Positives = 52/74 (70%), Gaps = 1/74 (1%)
Query 2 EDVAAIRSPFAATMMEGLPVVKQKKLEDYFP-DAPPMALDLLRQLLQFNPLKRISAEKAL 60
E++ + S ++A +++ L Q+ L+ P D PP ALDLL++LL F P KR+SAE+AL
Sbjct 241 EELQDLGSDYSALILQNLGSRPQQTLDALLPPDTPPEALDLLKRLLAFAPDKRLSAEQAL 300
Query 61 EHPYVRQFHNPEDE 74
+HPYV++FH P+ E
Sbjct 301 QHPYVQRFHCPDRE 314
> xla:397785 mapk1-b, Xp42, erk, erk2, mapk, mapk1b, mpk1; mitogen-activated
protein kinase 1 (EC:2.7.11.24); K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=361
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 34/76 (44%), Positives = 46/76 (60%), Gaps = 0/76 (0%)
Query 1 REDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKAL 60
+ED+ I + A + LP + FP+A P ALDLL ++L FNP KRI E AL
Sbjct 252 QEDLNCIINLKARNYLLSLPHKNKVPWNRLFPNADPKALDLLDKMLTFNPHKRIEVEAAL 311
Query 61 EHPYVRQFHNPEDEPV 76
HPY+ Q+++P DEPV
Sbjct 312 AHPYLEQYYDPSDEPV 327
> xla:398985 mapk1-a, MGC68934, erk, erk2, mapk, mapk1, mapk1a,
mpk1, xp42; mitogen-activated protein kinase 1 (EC:2.7.11.24);
K04371 extracellular signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=361
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 34/76 (44%), Positives = 46/76 (60%), Gaps = 0/76 (0%)
Query 1 REDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKAL 60
+ED+ I + A + LP + FP+A P ALDLL ++L FNP KRI E AL
Sbjct 252 QEDLNCIINLKARNYLLSLPHKNKVPWNRLFPNADPKALDLLDKMLTFNPHKRIEVEAAL 311
Query 61 EHPYVRQFHNPEDEPV 76
HPY+ Q+++P DEPV
Sbjct 312 AHPYLEQYYDPSDEPV 327
> dre:399480 mapk3, ERK1, fi06b09, wu:fi06b09, zERK1; mitogen-activated
protein kinase 3 (EC:2.7.11.1); K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=392
Score = 65.5 bits (158), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 33/76 (43%), Positives = 48/76 (63%), Gaps = 0/76 (0%)
Query 1 REDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKAL 60
++D+ I + A ++ LP + FP A ALDLL ++L FNPLKRI+ E+AL
Sbjct 280 QDDLNCIINMKARNYLQSLPQKPKIPWNKLFPKADNKALDLLDRMLTFNPLKRINVEQAL 339
Query 61 EHPYVRQFHNPEDEPV 76
HPY+ Q+++P DEPV
Sbjct 340 AHPYLEQYYDPSDEPV 355
> dre:100330818 mitogen-activated protein kinase 3-like; K04371
extracellular signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=392
Score = 65.5 bits (158), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 33/76 (43%), Positives = 48/76 (63%), Gaps = 0/76 (0%)
Query 1 REDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKAL 60
++D+ I + A ++ LP + FP A ALDLL ++L FNPLKRI+ E+AL
Sbjct 280 QDDLNCIINMKARNYLQSLPQKPKIPWNKLFPKADNKALDLLDRMLTFNPLKRINVEQAL 339
Query 61 EHPYVRQFHNPEDEPV 76
HPY+ Q+++P DEPV
Sbjct 340 AHPYLEQYYDPSDEPV 355
> mmu:26417 Mapk3, Erk-1, Erk1, Ert2, Esrk1, Mnk1, Mtap2k, Prkm3,
p44, p44erk1, p44mapk; mitogen-activated protein kinase
3 (EC:2.7.11.24); K04371 extracellular signal-regulated kinase
1/2 [EC:2.7.11.24]
Length=380
Score = 64.7 bits (156), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 32/76 (42%), Positives = 47/76 (61%), Gaps = 0/76 (0%)
Query 1 REDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKAL 60
+ED+ I + A ++ LP + FP + ALDLL ++L FNP KRI+ E+AL
Sbjct 267 QEDLNCIINMKARNYLQSLPSKTKVAWAKLFPKSDSKALDLLDRMLTFNPNKRITVEEAL 326
Query 61 EHPYVRQFHNPEDEPV 76
HPY+ Q+++P DEPV
Sbjct 327 AHPYLEQYYDPTDEPV 342
> hsa:225689 MAPK15, ERK7, ERK8; mitogen-activated protein kinase
15 (EC:2.7.11.24); K08293 mitogen-activated protein kinase
[EC:2.7.11.24]
Length=544
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 35/74 (47%), Positives = 49/74 (66%), Gaps = 1/74 (1%)
Query 2 EDVAAIRSPFAATMMEGLPVVKQKKLEDYFP-DAPPMALDLLRQLLQFNPLKRISAEKAL 60
ED+ A+ S A+++ L ++ L+ P D P ALDLLR+LL F P KR+SA +AL
Sbjct 240 EDLLALGSGCRASVLHQLGSRPRQTLDALLPPDTSPEALDLLRRLLVFAPDKRLSATQAL 299
Query 61 EHPYVRQFHNPEDE 74
+HPYV++FH P DE
Sbjct 300 QHPYVQRFHCPSDE 313
> hsa:5600 MAPK11, P38B, P38BETA2, PRKM11, SAPK2, SAPK2B, p38-2,
p38Beta; mitogen-activated protein kinase 11 (EC:2.7.11.24);
K04441 p38 MAP kinase [EC:2.7.11.24]
Length=364
Score = 64.7 bits (156), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 33/74 (44%), Positives = 47/74 (63%), Gaps = 0/74 (0%)
Query 2 EDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALE 61
E +A I S A T ++ LP + QK L F A P+A+DLL ++L + +R+SA +AL
Sbjct 245 EVLAKISSEHARTYIQSLPPMPQKDLSSIFRGANPLAIDLLGRMLVLDSDQRVSAAEALA 304
Query 62 HPYVRQFHNPEDEP 75
H Y Q+H+PEDEP
Sbjct 305 HAYFSQYHDPEDEP 318
> dre:541323 mapk7, bmk1, erk5, wu:fb73b02, zgc:113111; mitogen-activated
protein kinase 7 (EC:2.7.1.-); K04464 mitogen-activated
protein kinase 7 [EC:2.7.11.24]
Length=862
Score = 64.3 bits (155), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 32/73 (43%), Positives = 48/73 (65%), Gaps = 0/73 (0%)
Query 4 VAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALEHP 63
V +I S + + LP + L +P A P AL+LL +L+F+P +RISA +ALEHP
Sbjct 312 VGSIGSDRVRSYVRSLPSKAPEPLAALYPQAEPSALNLLAAMLRFDPRERISACQALEHP 371
Query 64 YVRQFHNPEDEPV 76
Y+ ++H+P+DEPV
Sbjct 372 YLSKYHDPDDEPV 384
> ath:AT3G59790 ATMPK10; MAP kinase/ kinase; K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=393
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 45/67 (67%), Gaps = 0/67 (0%)
Query 9 SPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALEHPYVRQF 68
S +A + LP + ++ + FP+ PP+A+DL+ ++L F+P +RIS ++AL HPY+ F
Sbjct 289 SEYAKRYIRQLPTLPRQSFTEKFPNVPPLAIDLVEKMLTFDPKQRISVKEALAHPYLSSF 348
Query 69 HNPEDEP 75
H+ DEP
Sbjct 349 HDITDEP 355
> dre:553780 mapk15, MGC110345, erk7, zgc:110345; mitogen-activated
protein kinase 15 (EC:2.7.11.24); K08293 mitogen-activated
protein kinase [EC:2.7.11.24]
Length=524
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 40/75 (53%), Positives = 54/75 (72%), Gaps = 1/75 (1%)
Query 2 EDVAAIRSPFAATMMEGLPVVKQKKLEDYFP-DAPPMALDLLRQLLQFNPLKRISAEKAL 60
EDV AIRS + A++++ + + Q L++ P PP ALDLL++LL FNP KR+SAE+AL
Sbjct 241 EDVLAIRSEYGASVIQRMLLRPQVPLDEILPASVPPDALDLLQRLLLFNPDKRLSAEEAL 300
Query 61 EHPYVRQFHNPEDEP 75
HPYV +FHNP EP
Sbjct 301 RHPYVSKFHNPSSEP 315
> mmu:26413 Mapk1, 9030612K14Rik, AA407128, AU018647, C78273,
ERK, Erk2, MAPK2, PRKM2, Prkm1, p41mapk, p42mapk; mitogen-activated
protein kinase 1 (EC:2.7.11.24); K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=358
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 32/76 (42%), Positives = 46/76 (60%), Gaps = 0/76 (0%)
Query 1 REDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKAL 60
+ED+ I + A + LP + FP+A ALDLL ++L FNP KRI E+AL
Sbjct 247 QEDLNCIINLKARNYLLSLPHKNKVPWNRLFPNADSKALDLLDKMLTFNPHKRIEVEQAL 306
Query 61 EHPYVRQFHNPEDEPV 76
HPY+ Q+++P DEP+
Sbjct 307 AHPYLEQYYDPSDEPI 322
> mmu:19094 Mapk11, DKFZp586C1322, P38b, Prkm11, Sapk2, Sapk2b,
p38-2, p38beta, p38beta2; mitogen-activated protein kinase
11 (EC:2.7.11.24); K04441 p38 MAP kinase [EC:2.7.11.24]
Length=364
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 32/74 (43%), Positives = 47/74 (63%), Gaps = 0/74 (0%)
Query 2 EDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALE 61
E +A I S A T ++ LP + QK L F A P+A+DLL ++L + +R+SA +AL
Sbjct 245 EVLAKISSEHARTYIQSLPPMPQKDLSSVFHGANPLAIDLLGRMLVLDSDQRVSAAEALA 304
Query 62 HPYVRQFHNPEDEP 75
H Y Q+H+P+DEP
Sbjct 305 HAYFSQYHDPDDEP 318
> hsa:5594 MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2,
p38, p40, p41, p41mapk; mitogen-activated protein kinase
1 (EC:2.7.11.24); K04371 extracellular signal-regulated kinase
1/2 [EC:2.7.11.24]
Length=360
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 32/76 (42%), Positives = 46/76 (60%), Gaps = 0/76 (0%)
Query 1 REDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKAL 60
+ED+ I + A + LP + FP+A ALDLL ++L FNP KRI E+AL
Sbjct 249 QEDLNCIINLKARNYLLSLPHKNKVPWNRLFPNADSKALDLLDKMLTFNPHKRIEVEQAL 308
Query 61 EHPYVRQFHNPEDEPV 76
HPY+ Q+++P DEP+
Sbjct 309 AHPYLEQYYDPSDEPI 324
> xla:494669 mapk11, p38-2, p38Beta, p38b, p38beta2, prkm11, sapk2,
sapk2b; mitogen-activated protein kinase 11; K04441 p38
MAP kinase [EC:2.7.11.24]
Length=361
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 31/69 (44%), Positives = 45/69 (65%), Gaps = 0/69 (0%)
Query 7 IRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALEHPYVR 66
I S A +E LP + + L++ F A P+A+DLL ++L + KRISA +AL HPY
Sbjct 249 ISSEHARRYIESLPYMPHQDLKEVFRGANPLAIDLLEKMLILDSDKRISATEALAHPYFV 308
Query 67 QFHNPEDEP 75
Q+H+P+DEP
Sbjct 309 QYHDPDDEP 317
> dre:562552 fc89f08, wu:fc89f08; zgc:171775; K04441 p38 MAP kinase
[EC:2.7.11.24]
Length=359
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 30/73 (41%), Positives = 46/73 (63%), Gaps = 0/73 (0%)
Query 4 VAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALEHP 63
V ++S A + + LPV K+K ++ F P A+DLL +L +P R+SA+ L HP
Sbjct 248 VLKMQSKDAQSYVRSLPVQKKKAFKEVFSGMDPNAIDLLEGMLVLDPEVRLSAKNGLSHP 307
Query 64 YVRQFHNPEDEPV 76
Y+ +FH+PE+EPV
Sbjct 308 YLSEFHDPENEPV 320
> dre:415185 zgc:86905; K04441 p38 MAP kinase [EC:2.7.11.24]
Length=361
Score = 61.6 bits (148), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 31/69 (44%), Positives = 44/69 (63%), Gaps = 0/69 (0%)
Query 7 IRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALEHPYVR 66
I S A ++ LP + Q+ L F A P+A+DLL+++L + RISA +AL HPY
Sbjct 249 ISSEHAQKYIQSLPHMPQQDLGKIFRGANPLAVDLLKKMLVLDCDGRISASEALCHPYFS 308
Query 67 QFHNPEDEP 75
Q+H+PEDEP
Sbjct 309 QYHDPEDEP 317
> hsa:5595 MAPK3, ERK1, HS44KDAP, HUMKER1A, MGC20180, P44ERK1,
P44MAPK, PRKM3; mitogen-activated protein kinase 3 (EC:2.7.11.24);
K04371 extracellular signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=357
Score = 61.2 bits (147), Expect = 7e-10, Method: Composition-based stats.
Identities = 30/74 (40%), Positives = 45/74 (60%), Gaps = 0/74 (0%)
Query 1 REDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKAL 60
+ED+ I + A ++ LP + FP + ALDLL ++L FNP KRI+ E+AL
Sbjct 266 QEDLNCIINMKARNYLQSLPSKTKVAWAKLFPKSDSKALDLLDRMLTFNPNKRITVEEAL 325
Query 61 EHPYVRQFHNPEDE 74
HPY+ Q+++P DE
Sbjct 326 AHPYLEQYYDPTDE 339
> xla:100329215 p38-beta; mitogen-activated protein kinase p38
beta; K04441 p38 MAP kinase [EC:2.7.11.24]
Length=361
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 30/69 (43%), Positives = 45/69 (65%), Gaps = 0/69 (0%)
Query 7 IRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALEHPYVR 66
I S A +E LP + Q+ L++ F A P+A+DLL ++L + KRI+A +AL H Y
Sbjct 249 ISSEHARRYIESLPYMPQQDLKEVFHGANPLAIDLLEKMLILDSDKRIAATEALAHAYFV 308
Query 67 QFHNPEDEP 75
Q+H+P+DEP
Sbjct 309 QYHDPDDEP 317
> cel:F43C1.2 mpk-1; MAP Kinase family member (mpk-1); K04371
extracellular signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=444
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 31/74 (41%), Positives = 45/74 (60%), Gaps = 0/74 (0%)
Query 3 DVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALEH 62
D+ I + A + + LP ++ +P A P ALDLL ++L FNP RI E+AL H
Sbjct 322 DLQCIINDKARSYLISLPHKPKQPWARLYPGADPRALDLLDKMLTFNPHNRIDIEQALAH 381
Query 63 PYVRQFHNPEDEPV 76
PY+ Q+++P DEPV
Sbjct 382 PYLEQYYDPGDEPV 395
> sce:YHR030C SLT2, BYC2, MPK1, SLK2; Serine/threonine MAP kinase
involved in regulating the maintenance of cell wall integrity
and progression through the cell cycle; regulated by the
PKC1-mediated signaling pathway (EC:2.7.11.24); K04464 mitogen-activated
protein kinase 7 [EC:2.7.11.24]
Length=484
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 26/46 (56%), Positives = 36/46 (78%), Gaps = 0/46 (0%)
Query 31 FPDAPPMALDLLRQLLQFNPLKRISAEKALEHPYVRQFHNPEDEPV 76
+P+A ALDLL Q+L F+P KRI+ ++ALEHPY+ +H+P DEPV
Sbjct 284 YPNANSQALDLLEQMLAFDPQKRITVDEALEHPYLSIWHDPADEPV 329
> xla:379992 mapk14, MGC69036, csbp, mapk14a, mxi2, p38, p38-alpha,
sapk2, sapk2a; mitogen-activated protein kinase 14 (EC:2.7.11.24);
K04441 p38 MAP kinase [EC:2.7.11.24]
Length=361
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 29/70 (41%), Positives = 43/70 (61%), Gaps = 0/70 (0%)
Query 7 IRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALEHPYVR 66
I S A ++ LP + + ED F A P A+DLL ++L + KRI+A +AL H Y
Sbjct 251 ISSEAARNYIQSLPYMPKMNFEDVFLGANPQAVDLLEKMLVLDTDKRITAAEALAHSYFA 310
Query 67 QFHNPEDEPV 76
Q+H+P+DEP+
Sbjct 311 QYHDPDDEPI 320
> dre:65237 mapk14a, MAPK14, fk28c03, hm:zeh1243, p38, p38a, wu:fk28c03,
zp38a; mitogen-activated protein kinase 14a (EC:2.7.11.24);
K04441 p38 MAP kinase [EC:2.7.11.24]
Length=361
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 28/64 (43%), Positives = 41/64 (64%), Gaps = 0/64 (0%)
Query 12 AATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALEHPYVRQFHNP 71
A T + LP + ++ D F A P A+DLL ++L + KRI+A +AL HPY Q+H+P
Sbjct 256 ARTYISSLPQMPKRNFADVFIGANPQAVDLLEKMLVLDTDKRITAAEALAHPYFAQYHDP 315
Query 72 EDEP 75
+DEP
Sbjct 316 DDEP 319
> dre:266756 mapk14b, MAPK14, p38b, zp38b; mitogen-activated protein
kinase 14b (EC:2.7.11.24); K04441 p38 MAP kinase [EC:2.7.11.24]
Length=348
Score = 58.9 bits (141), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 27/64 (42%), Positives = 41/64 (64%), Gaps = 0/64 (0%)
Query 12 AATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALEHPYVRQFHNP 71
A + LP + ++ D F A P+A+DLL ++L + KRI+A +AL HPY Q+H+P
Sbjct 256 ARNYISSLPHMPKRNFADVFIGANPLAVDLLEKMLVLDTDKRITASQALAHPYFAQYHDP 315
Query 72 EDEP 75
+DEP
Sbjct 316 DDEP 319
> ath:AT2G18170 ATMPK7; ATMPK7 (ARABIDOPSIS THALIANA MAP KINASE
7); MAP kinase/ kinase; K08293 mitogen-activated protein kinase
[EC:2.7.11.24]
Length=368
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 28/74 (37%), Positives = 43/74 (58%), Gaps = 0/74 (0%)
Query 2 EDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALE 61
D+ I +P A ++ LP + L + +P A P+A+DLL+++L F+P KRIS AL
Sbjct 256 SDIRFIDNPKARRFIKSLPYSRGTHLSNLYPQANPLAIDLLQRMLVFDPTKRISVTDALL 315
Query 62 HPYVRQFHNPEDEP 75
HPY+ +P P
Sbjct 316 HPYMAGLFDPGSNP 329
> ath:AT1G07880 ATMPK13; MAP kinase/ kinase; K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=363
Score = 57.8 bits (138), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 28/74 (37%), Positives = 46/74 (62%), Gaps = 0/74 (0%)
Query 3 DVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALEH 62
D+ +RS A ++ LP V+++ + FP+ PMALDL ++L F+P KRI+ ++AL+
Sbjct 257 DLDFLRSDNARKYVKQLPHVQKQSFREKFPNISPMALDLAEKMLVFDPSKRITVDEALKQ 316
Query 63 PYVRQFHNPEDEPV 76
PY+ H +EP
Sbjct 317 PYLASLHEINEEPT 330
> sce:YGR040W KSS1; Kss1p (EC:2.7.11.24); K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=368
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 33/76 (43%), Positives = 44/76 (57%), Gaps = 2/76 (2%)
Query 2 EDVAAIRSPFAATMMEGLPVVKQKKLEDYFP--DAPPMALDLLRQLLQFNPLKRISAEKA 59
ED I+S A + LP+ E + D P +DLL ++LQFNP KRISA +A
Sbjct 248 EDFNQIKSKRAKEYIANLPMRPPLPWETVWSKTDLNPDMIDLLDKMLQFNPDKRISAAEA 307
Query 60 LEHPYVRQFHNPEDEP 75
L HPY+ +H+P DEP
Sbjct 308 LRHPYLAMYHDPSDEP 323
> hsa:6300 MAPK12, ERK3, ERK6, P38GAMMA, PRKM12, SAPK-3, SAPK3;
mitogen-activated protein kinase 12 (EC:2.7.11.24); K04441
p38 MAP kinase [EC:2.7.11.24]
Length=367
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 45/74 (60%), Gaps = 0/74 (0%)
Query 2 EDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALE 61
E V ++S A M+GLP +++K +A P+A++LL ++L + +R++A +AL
Sbjct 248 EFVQRLQSDEAKNYMKGLPELEKKDFASILTNASPLAVNLLEKMLVLDAEQRVTAGEALA 307
Query 62 HPYVRQFHNPEDEP 75
HPY H+ EDEP
Sbjct 308 HPYFESLHDTEDEP 321
> hsa:100509672 mitogen-activated protein kinase 12-like
Length=270
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 45/74 (60%), Gaps = 0/74 (0%)
Query 2 EDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALE 61
E V ++S A M+GLP +++K +A P+A++LL ++L + +R++A +AL
Sbjct 151 EFVQRLQSDEAKNYMKGLPELEKKDFASILTNASPLAVNLLEKMLVLDAEQRVTAGEALA 210
Query 62 HPYVRQFHNPEDEP 75
HPY H+ EDEP
Sbjct 211 HPYFESLHDTEDEP 224
> sce:YKL161C MLP1; Protein kinase implicated in the Slt2p mitogen-activated
(MAP) kinase signaling pathway; associates with
Rlm1p (EC:2.7.1.-); K08293 mitogen-activated protein kinase
[EC:2.7.11.24]
Length=433
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 23/50 (46%), Positives = 36/50 (72%), Gaps = 0/50 (0%)
Query 25 KKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALEHPYVRQFHNPEDE 74
+ E P A P AL+LL+++L+F+P KRI+ E ALEHPY+ +H+ ++E
Sbjct 278 RSFESILPGANPEALELLKKMLEFDPKKRITVEDALEHPYLSMWHDIDEE 327
> hsa:100509694 mitogen-activated protein kinase 12-like
Length=357
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 45/74 (60%), Gaps = 0/74 (0%)
Query 2 EDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALE 61
E V ++S A M+GLP +++K +A P+A++LL ++L + +R++A +AL
Sbjct 238 EFVQRLQSDEAKNYMKGLPELEKKDFASILTNASPLAVNLLEKMLVLDAEQRVTAGEALA 297
Query 62 HPYVRQFHNPEDEP 75
HPY H+ EDEP
Sbjct 298 HPYFESLHDTEDEP 311
> bbo:BBOV_IV005520 23.m06268; mitogen-activated protein kinase;
K08293 mitogen-activated protein kinase [EC:2.7.11.24]
Length=584
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 29/71 (40%), Positives = 41/71 (57%), Gaps = 0/71 (0%)
Query 2 EDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALE 61
ED+AAI P + K L F PP A++LL+Q+L FNP KRI+ +AL+
Sbjct 465 EDIAAIIKPEVRRYIRMFHPRKGIDLYKRFKGTPPAAVNLLQQMLTFNPYKRITVAEALK 524
Query 62 HPYVRQFHNPE 72
H Y R+F+ P+
Sbjct 525 HEYFREFYKPQ 535
> ath:AT4G36450 ATMPK14 (Mitogen-activated protein kinase 14);
MAP kinase/ kinase; K04371 extracellular signal-regulated kinase
1/2 [EC:2.7.11.24]
Length=361
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 28/73 (38%), Positives = 40/73 (54%), Gaps = 0/73 (0%)
Query 3 DVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALEH 62
D+ I + A ++ LP K +P A P+A+DLL+++L F+P KRIS AL H
Sbjct 254 DLQFIDNQKARRFIKSLPFSKGTHFSHIYPHANPLAIDLLQRMLVFDPTKRISVSDALLH 313
Query 63 PYVRQFHNPEDEP 75
PY+ PE P
Sbjct 314 PYMEGLLEPECNP 326
> cel:B0218.3 pmk-1; P38 Map Kinase family member (pmk-1); K04441
p38 MAP kinase [EC:2.7.11.24]
Length=377
Score = 54.7 bits (130), Expect = 7e-08, Method: Composition-based stats.
Identities = 25/75 (33%), Positives = 44/75 (58%), Gaps = 0/75 (0%)
Query 2 EDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALE 61
E + I S A + LP + ++ + F A P A+DLL ++L +P +R +A++A+E
Sbjct 256 EFLKKISSEEARNYIRNLPKMTRRDFKRLFAQATPQAIDLLEKMLHLDPDRRPTAKEAME 315
Query 62 HPYVRQFHNPEDEPV 76
H Y+ +H+ DEP+
Sbjct 316 HEYLAAYHDETDEPI 330
> ath:AT3G45640 ATMPK3; ATMPK3 (ARABIDOPSIS THALIANA MITOGEN-ACTIVATED
PROTEIN KINASE 3); MAP kinase/ kinase/ protein binding
/ protein kinase; K04371 extracellular signal-regulated
kinase 1/2 [EC:2.7.11.24]
Length=370
Score = 54.7 bits (130), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 26/75 (34%), Positives = 43/75 (57%), Gaps = 0/75 (0%)
Query 2 EDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALE 61
D+ + A + LP ++ L F PMA+DL+ ++L F+P +RI+ E+AL
Sbjct 261 SDLGFTHNEDAKRYIRQLPNFPRQPLAKLFSHVNPMAIDLVDRMLTFDPNRRITVEQALN 320
Query 62 HPYVRQFHNPEDEPV 76
H Y+ + H+P DEP+
Sbjct 321 HQYLAKLHDPNDEPI 335
> mmu:29857 Mapk12, AW123708, Erk6, P38gamma, Prkm12, Sapk3; mitogen-activated
protein kinase 12 (EC:2.7.11.24); K04441 p38
MAP kinase [EC:2.7.11.24]
Length=367
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 43/74 (58%), Gaps = 0/74 (0%)
Query 2 EDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALE 61
E V ++S A MEGLP +++K +A P A++LL ++L + +R++A +AL
Sbjct 248 EFVQKLQSAEAKNYMEGLPELEKKDFASVLTNASPQAVNLLERMLVLDAEQRVTAAEALT 307
Query 62 HPYVRQFHNPEDEP 75
HPY + EDEP
Sbjct 308 HPYFESLRDTEDEP 321
> ath:AT4G11330 ATMPK5; ATMPK5 (MAP KINASE 5); MAP kinase/ kinase;
K04371 extracellular signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=376
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 27/70 (38%), Positives = 42/70 (60%), Gaps = 0/70 (0%)
Query 7 IRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALEHPYVR 66
+RS A ++ LP ++ FP A+DLL ++L F+P+KRI+ E+AL +PY+
Sbjct 271 LRSANARKYVKELPKFPRQNFSARFPSMNSTAIDLLEKMLVFDPVKRITVEEALCYPYLS 330
Query 67 QFHNPEDEPV 76
H+ DEPV
Sbjct 331 ALHDLNDEPV 340
> ath:AT1G10210 ATMPK1; ATMPK1 (MITOGEN-ACTIVATED PROTEIN KINASE
1); MAP kinase/ kinase; K04371 extracellular signal-regulated
kinase 1/2 [EC:2.7.11.24]
Length=370
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 28/74 (37%), Positives = 43/74 (58%), Gaps = 0/74 (0%)
Query 2 EDVAAIRSPFAATMMEGLPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALE 61
ED+ I +P A + LP L +P A +A+DLL+++L F+P KRIS +AL+
Sbjct 256 EDLEFIDNPKAKRYIRSLPYSPGMSLSRLYPGAHVLAIDLLQKMLVFDPSKRISVSEALQ 315
Query 62 HPYVRQFHNPEDEP 75
HPY+ ++P P
Sbjct 316 HPYMAPLYDPNANP 329
> sce:YLR113W HOG1, SSK3; Hog1p (EC:2.7.11.24); K04441 p38 MAP
kinase [EC:2.7.11.24]
Length=435
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 24/50 (48%), Positives = 32/50 (64%), Gaps = 0/50 (0%)
Query 27 LEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALEHPYVRQFHNPEDEPV 76
+ F P A+DLL ++L F+P KRI+A AL HPY +H+P DEPV
Sbjct 264 FSERFKTVEPDAVDLLEKMLVFDPKKRITAADALAHPYSAPYHDPTDEPV 313
> ath:AT2G43790 ATMPK6; ATMPK6 (ARABIDOPSIS THALIANA MAP KINASE
6); MAP kinase/ kinase; K14512 mitogen-activated protein kinase
6 [EC:2.7.11.24]
Length=395
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 23/57 (40%), Positives = 36/57 (63%), Gaps = 0/57 (0%)
Query 19 LPVVKQKKLEDYFPDAPPMALDLLRQLLQFNPLKRISAEKALEHPYVRQFHNPEDEP 75
LP ++ + D FP P+A+DL+ ++L F+P +RI+ AL HPY+ H+ DEP
Sbjct 302 LPPYPRQSITDKFPTVHPLAIDLIEKMLTFDPRRRITVLDALAHPYLNSLHDISDEP 358
Lambda K H
0.319 0.137 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2076916240
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40