bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5827_orf2
Length=109
Score E
Sequences producing significant alignments: (Bits) Value
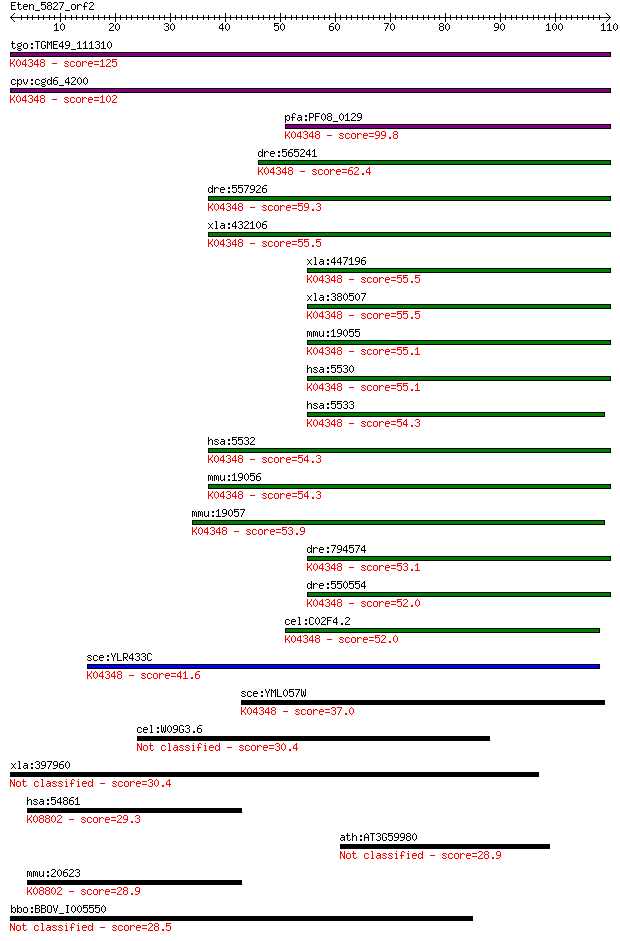
tgo:TGME49_111310 serine/threonine protein phosphatase, putati... 125 3e-29
cpv:cgd6_4200 protein phosphatase 2B catalytic subunit, calcin... 102 2e-22
pfa:PF08_0129 serine/threonine protein phosphatase, putative; ... 99.8 2e-21
dre:565241 ppp3ccb, im:7163784, zgc:158685; protein phosphatas... 62.4 3e-10
dre:557926 ppp3cca, ppp3cc, sb:cb767, si:dkey-21k4.5; protein ... 59.3 3e-09
xla:432106 ppp3cb, MGC81043, calna2, calnb, cna2, pp2bbeta; pr... 55.5 4e-08
xla:447196 ppp3ca, MGC79144, calcineurin, caln, calna, calna1,... 55.5 5e-08
xla:380507 ppp3ca, MGC52521; protein phosphatase 3 (formerly 2... 55.5 5e-08
mmu:19055 Ppp3ca, 2900074D19Rik, AI841391, AW413465, CN, Caln,... 55.1 5e-08
hsa:5530 PPP3CA, CALN, CALNA, CALNA1, CCN1, CNA1, PPP2B; prote... 55.1 5e-08
hsa:5533 PPP3CC, CALNA3, CNA3, PP2Bgamma; protein phosphatase ... 54.3 9e-08
hsa:5532 PPP3CB, CALNA2, CALNB, CNA2, PP2Bbeta; protein phosph... 54.3 9e-08
mmu:19056 Ppp3cb, 1110063J16Rik, Calnb, CnAbeta, Cnab; protein... 54.3 9e-08
mmu:19057 Ppp3cc, Calnc; protein phosphatase 3, catalytic subu... 53.9 1e-07
dre:794574 ppp3ca, fj46e08, si:dkeyp-79c2.2, wu:fj46e08; prote... 53.1 2e-07
dre:550554 ppp3cb, wu:fq19c08, zgc:110141; protein phosphatase... 52.0 4e-07
cel:C02F4.2 tax-6; abnormal CHEmotaxis family member (tax-6); ... 52.0 5e-07
sce:YLR433C CNA1, CMP1; Calcineurin A; one isoform (the other ... 41.6 7e-04
sce:YML057W CMP2, CNA2; Calcineurin A; one isoform (the other ... 37.0 0.014
cel:W09G3.6 hypothetical protein 30.4 1.5
xla:397960 myf5, Xmyf5, xmyf-5; myogenic factor 5 30.4
hsa:54861 SNRK, DKFZp779A1866, FLJ20224, HSNFRK, KIAA0096; SNF... 29.3 3.4
ath:AT3G59980 tRNA-binding region domain-containing protein 28.9 4.3
mmu:20623 Snrk, 2010012F07Rik, AI448042, AW547029, E030034B15,... 28.9 4.5
bbo:BBOV_I005550 26.m00079; variant erythrocyte surface antigen-1 28.5 5.5
> tgo:TGME49_111310 serine/threonine protein phosphatase, putative
(EC:3.1.3.16); K04348 protein phosphatase 3, catalytic
subunit [EC:3.1.3.16]
Length=501
Score = 125 bits (315), Expect = 3e-29, Method: Composition-based stats.
Identities = 70/111 (63%), Positives = 92/111 (82%), Gaps = 2/111 (1%)
Query 1 LYGILNPSLEEGEEDEEIDNVELPPEVVKIMRANIVLAEEGTA--PRSAGTPISRERAEA 58
LYGILNPS+++ E+DE++D+VELPP V+ IM+A++ E P + +S+ERA+A
Sbjct 365 LYGILNPSIDDDEDDEDVDDVELPPAVLSIMKAHLPSDEASGQRHPPAGDNRLSKERADA 424
Query 59 LRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVGLLIQGREGLEN 109
LR+KVQSVGRLMRVFKT+R+ENELI++LKGCTPG RIPVGLL+QGREG+ N
Sbjct 425 LRKKVQSVGRLMRVFKTLRQENELIVRLKGCTPGHRIPVGLLLQGREGIAN 475
> cpv:cgd6_4200 protein phosphatase 2B catalytic subunit, calcineurin
like phosphatse superfamily ; K04348 protein phosphatase
3, catalytic subunit [EC:3.1.3.16]
Length=504
Score = 102 bits (255), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 56/123 (45%), Positives = 78/123 (63%), Gaps = 15/123 (12%)
Query 1 LYGILNPSLEEGEEDEEIDNVELPPEVVKIMRANIVLAEEGTAP--------------RS 46
LY I+ P E E++ +LP +V ++M A + E+G P +
Sbjct 363 LYSIIQPVATSSSEPMELEEEDLPDDVRRLMDA-FCINEDGKLPEEVASLGSSIIRSAQP 421
Query 47 AGTPISRERAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVGLLIQGREG 106
A + IS+ERA+ LR+KVQSV R+MRVF T+RE+NELI++LKG TPG RIP+GLL+ GR+
Sbjct 422 ASSVISKERADTLRKKVQSVARIMRVFSTLREQNELIVKLKGVTPGHRIPMGLLLGGRDA 481
Query 107 LEN 109
LEN
Sbjct 482 LEN 484
> pfa:PF08_0129 serine/threonine protein phosphatase, putative;
K04348 protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=604
Score = 99.8 bits (247), Expect = 2e-21, Method: Composition-based stats.
Identities = 44/59 (74%), Positives = 55/59 (93%), Gaps = 0/59 (0%)
Query 51 ISRERAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVGLLIQGREGLEN 109
+S+ER + LR+KVQSVGRLMRVF+T+R+ENELI+QLKGC+PG RIPVGLL+ G+EGLEN
Sbjct 524 VSKERTDTLRKKVQSVGRLMRVFRTLRKENELIVQLKGCSPGYRIPVGLLLSGKEGLEN 582
> dre:565241 ppp3ccb, im:7163784, zgc:158685; protein phosphatase
3, catalytic subunit, gamma isozyme, b (EC:3.1.3.16); K04348
protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=506
Score = 62.4 bits (150), Expect = 3e-10, Method: Composition-based stats.
Identities = 27/64 (42%), Positives = 43/64 (67%), Gaps = 0/64 (0%)
Query 46 SAGTPISRERAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVGLLIQGRE 105
S G R E +R K++++G++ RVF +REENE ++QLKG TP +P+G+L GR+
Sbjct 376 SEGDDTCEVRKEVIRNKIRAIGKMARVFSVLREENESVLQLKGLTPTGMLPLGVLSGGRQ 435
Query 106 GLEN 109
L++
Sbjct 436 TLQS 439
> dre:557926 ppp3cca, ppp3cc, sb:cb767, si:dkey-21k4.5; protein
phosphatase 3, catalytic subunit, gamma isozyme, a (EC:3.1.3.16);
K04348 protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=499
Score = 59.3 bits (142), Expect = 3e-09, Method: Composition-based stats.
Identities = 28/73 (38%), Positives = 46/73 (63%), Gaps = 2/73 (2%)
Query 37 LAEEGTAPRSAGTPISRERAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIP 96
L EG G P R+ E +R K++++G++ RVF +REE+E ++ LKG TP +P
Sbjct 375 LMSEGDDLCEGGAPAVRK--EVIRNKIRAIGKMARVFSVLREESESVLTLKGLTPTGTLP 432
Query 97 VGLLIQGREGLEN 109
+G+L G++ L+N
Sbjct 433 LGVLSGGKQTLQN 445
> xla:432106 ppp3cb, MGC81043, calna2, calnb, cna2, pp2bbeta;
protein phosphatase 3, catalytic subunit, beta isozyme; K04348
protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=507
Score = 55.5 bits (132), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 46/73 (63%), Gaps = 3/73 (4%)
Query 37 LAEEGTAPRSAGTPISRERAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIP 96
L +G P +G + R E +R K++++G++ RVF +REE+E ++ LKG TP +P
Sbjct 375 LITDGEDPLESGASV---RKEIIRNKIRAIGKMARVFSVLREESESVLTLKGLTPTGTLP 431
Query 97 VGLLIQGREGLEN 109
G+L GR+ L++
Sbjct 432 QGVLAGGRQTLQS 444
> xla:447196 ppp3ca, MGC79144, calcineurin, caln, calna, calna1,
ccn1, cna1, ppp2b; protein phosphatase 3, catalytic subunit,
alpha isozyme; K04348 protein phosphatase 3, catalytic subunit
[EC:3.1.3.16]
Length=511
Score = 55.5 bits (132), Expect = 5e-08, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 39/55 (70%), Gaps = 0/55 (0%)
Query 55 RAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVGLLIQGREGLEN 109
R E +R K++++G++ RVF +REE+E ++ LKG TP +P G+L G++ L++
Sbjct 392 RKEVIRNKIRAIGKMARVFSVLREESESVLTLKGLTPTGMLPSGVLSGGKQTLQS 446
> xla:380507 ppp3ca, MGC52521; protein phosphatase 3 (formerly
2B), catalytic subunit, alpha isoform (calcineurin A alpha)
(EC:3.1.3.16); K04348 protein phosphatase 3, catalytic subunit
[EC:3.1.3.16]
Length=518
Score = 55.5 bits (132), Expect = 5e-08, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 39/55 (70%), Gaps = 0/55 (0%)
Query 55 RAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVGLLIQGREGLEN 109
R E +R K++++G++ RVF +REE+E ++ LKG TP +P G+L G++ L++
Sbjct 392 RKEVIRNKIRAIGKMARVFSVLREESESVLTLKGLTPTGMLPSGVLSGGKQTLQS 446
> mmu:19055 Ppp3ca, 2900074D19Rik, AI841391, AW413465, CN, Caln,
Calna, CnA, MGC106804; protein phosphatase 3, catalytic subunit,
alpha isoform (EC:3.1.3.16); K04348 protein phosphatase
3, catalytic subunit [EC:3.1.3.16]
Length=521
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 39/55 (70%), Gaps = 0/55 (0%)
Query 55 RAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVGLLIQGREGLEN 109
R E +R K++++G++ RVF +REE+E ++ LKG TP +P G+L G++ L++
Sbjct 392 RKEVIRNKIRAIGKMARVFSVLREESESVLTLKGLTPTGMLPSGVLSGGKQTLQS 446
> hsa:5530 PPP3CA, CALN, CALNA, CALNA1, CCN1, CNA1, PPP2B; protein
phosphatase 3, catalytic subunit, alpha isozyme (EC:3.1.3.16);
K04348 protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=521
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 39/55 (70%), Gaps = 0/55 (0%)
Query 55 RAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVGLLIQGREGLEN 109
R E +R K++++G++ RVF +REE+E ++ LKG TP +P G+L G++ L++
Sbjct 392 RKEVIRNKIRAIGKMARVFSVLREESESVLTLKGLTPTGMLPSGVLSGGKQTLQS 446
> hsa:5533 PPP3CC, CALNA3, CNA3, PP2Bgamma; protein phosphatase
3, catalytic subunit, gamma isozyme (EC:3.1.3.16); K04348
protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=512
Score = 54.3 bits (129), Expect = 9e-08, Method: Composition-based stats.
Identities = 21/54 (38%), Positives = 39/54 (72%), Gaps = 0/54 (0%)
Query 55 RAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVGLLIQGREGLE 108
R E +R K++++G++ RVF +R+E+E ++ LKG TP +P+G+L G++ +E
Sbjct 386 RKEIIRNKIRAIGKMARVFSILRQESESVLTLKGLTPTGTLPLGVLSGGKQTIE 439
> hsa:5532 PPP3CB, CALNA2, CALNB, CNA2, PP2Bbeta; protein phosphatase
3, catalytic subunit, beta isozyme (EC:3.1.3.16); K04348
protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=525
Score = 54.3 bits (129), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 46/73 (63%), Gaps = 2/73 (2%)
Query 37 LAEEGTAPRSAGTPISRERAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIP 96
L EG G+ +R+ E +R K++++G++ RVF +REE+E ++ LKG TP +P
Sbjct 386 LMTEGEDQFDVGSAAARK--EIIRNKIRAIGKMARVFSVLREESESVLTLKGLTPTGMLP 443
Query 97 VGLLIQGREGLEN 109
G+L GR+ L++
Sbjct 444 SGVLAGGRQTLQS 456
> mmu:19056 Ppp3cb, 1110063J16Rik, Calnb, CnAbeta, Cnab; protein
phosphatase 3, catalytic subunit, beta isoform (EC:3.1.3.16);
K04348 protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=525
Score = 54.3 bits (129), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 46/73 (63%), Gaps = 2/73 (2%)
Query 37 LAEEGTAPRSAGTPISRERAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIP 96
L EG G+ +R+ E +R K++++G++ RVF +REE+E ++ LKG TP +P
Sbjct 386 LMTEGEDQFDVGSAAARK--EIIRNKIRAIGKMARVFSVLREESESVLTLKGLTPTGMLP 443
Query 97 VGLLIQGREGLEN 109
G+L GR+ L++
Sbjct 444 SGVLAGGRQTLQS 456
> mmu:19057 Ppp3cc, Calnc; protein phosphatase 3, catalytic subunit,
gamma isoform (EC:3.1.3.16); K04348 protein phosphatase
3, catalytic subunit [EC:3.1.3.16]
Length=513
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 26/75 (34%), Positives = 45/75 (60%), Gaps = 2/75 (2%)
Query 34 NIVLAEEGTAPRSAGTPISRERAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGD 93
NI EE G R+ E ++ K++++G++ RVF +REE+E ++ LKG TP
Sbjct 366 NICSDEEMNVTDEEGATTGRK--EVIKNKIRAIGKMARVFTVLREESENVLTLKGLTPTG 423
Query 94 RIPVGLLIQGREGLE 108
+P+G+L G++ +E
Sbjct 424 TLPLGVLSGGKQTIE 438
> dre:794574 ppp3ca, fj46e08, si:dkeyp-79c2.2, wu:fj46e08; protein
phosphatase 3, catalytic subunit, alpha isozyme; K04348
protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=521
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 20/55 (36%), Positives = 39/55 (70%), Gaps = 0/55 (0%)
Query 55 RAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVGLLIQGREGLEN 109
R E +R K++++G++ ++F +REE+E ++ LKG TP +P G+L G++ L++
Sbjct 392 RKEVIRNKIRAIGKMAKMFSVLREESENVLTLKGLTPTGMLPSGVLAGGKQTLQS 446
> dre:550554 ppp3cb, wu:fq19c08, zgc:110141; protein phosphatase
3, catalytic subunit, beta isozyme (EC:3.1.3.16); K04348
protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=511
Score = 52.0 bits (123), Expect = 4e-07, Method: Composition-based stats.
Identities = 20/55 (36%), Positives = 39/55 (70%), Gaps = 0/55 (0%)
Query 55 RAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVGLLIQGREGLEN 109
R E +R K++++G++ ++F +REE+E ++ LKG TP +P G+L G++ L++
Sbjct 382 RKEVIRNKIRAIGKMAKMFSVLREESENVLTLKGLTPTGMLPSGVLSGGKQTLQS 436
> cel:C02F4.2 tax-6; abnormal CHEmotaxis family member (tax-6);
K04348 protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=535
Score = 52.0 bits (123), Expect = 5e-07, Method: Composition-based stats.
Identities = 21/57 (36%), Positives = 36/57 (63%), Gaps = 0/57 (0%)
Query 51 ISRERAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVGLLIQGREGL 107
+ R E +R K++++G++ R F +REE+E ++ LKG TP +P+G L G G+
Sbjct 414 VGSARKEVIRHKIRAIGKMARAFSVLREESESVLALKGLTPTGALPMGTLQGGSRGV 470
> sce:YLR433C CNA1, CMP1; Calcineurin A; one isoform (the other
is CMP2) of the catalytic subunit of calcineurin, a Ca++/calmodulin-regulated
protein phosphatase which regulates Crz1p
(a stress-response transcription factor), the other calcineurin
subunit is CNB1 (EC:3.1.3.16); K04348 protein phosphatase
3, catalytic subunit [EC:3.1.3.16]
Length=553
Score = 41.6 bits (96), Expect = 7e-04, Method: Composition-based stats.
Identities = 28/97 (28%), Positives = 48/97 (49%), Gaps = 5/97 (5%)
Query 15 DEEIDNVELPPEVVKIMRANIVLAEEGTAPR----SAGTPISRERAEALRRKVQSVGRLM 70
++E+D P + ++A +E P S+ R +ALR K+ ++ ++
Sbjct 410 EQELDPESEPKAAEETVKARANATKETGTPSDEKASSAILEDETRRKALRNKILAIAKVS 469
Query 71 RVFKTIREENELIMQLKGCTPGDRIPVGLLIQGREGL 107
R+F +REE+E + LK G +P G L +G EGL
Sbjct 470 RMFSVLREESEKVEYLKTMNAG-VLPRGALARGTEGL 505
> sce:YML057W CMP2, CNA2; Calcineurin A; one isoform (the other
is CNA1) of the catalytic subunit of calcineurin, a Ca++/calmodulin-regulated
protein phosphatase which regulates Crz1p
(a stress-response transcription factor), the other calcineurin
subunit is CNB1 (EC:3.1.3.16); K04348 protein phosphatase
3, catalytic subunit [EC:3.1.3.16]
Length=604
Score = 37.0 bits (84), Expect = 0.014, Method: Composition-based stats.
Identities = 21/66 (31%), Positives = 35/66 (53%), Gaps = 1/66 (1%)
Query 43 APRSAGTPISRERAEALRRKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVGLLIQ 102
+P+ A R +ALR K+ +V ++ R++ +REE + LK G +P G L
Sbjct 489 SPKHASILDDEHRRKALRNKILAVAKVSRMYSVLREETNKVQFLKDHNSG-VLPRGALSN 547
Query 103 GREGLE 108
G +GL+
Sbjct 548 GVKGLD 553
> cel:W09G3.6 hypothetical protein
Length=1125
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 34/64 (53%), Gaps = 2/64 (3%)
Query 24 PPEVVKIMRANIVLAEEGTAPRSAGTPISRERAEALRRKVQSVGRLMRVFKTIREENELI 83
PP++VK++R L T P + TPI+ A A V ++G + + R+++E+
Sbjct 644 PPQIVKVIRPGHRLQTTTTKPIATVTPITVPVANADDVAVNNMGMISEEMR--RKQDEMW 701
Query 84 MQLK 87
+ L+
Sbjct 702 LDLR 705
> xla:397960 myf5, Xmyf5, xmyf-5; myogenic factor 5
Length=255
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 40/99 (40%), Gaps = 20/99 (20%)
Query 1 LYGILNPSLEEGEEDEEIDNVELPPEVVKIMRANIVLAEEGTAPRSAGTPISRERAEALR 60
+YG LEE +EDE + RA I + G A R+ + R
Sbjct 38 IYGAHKKDLEESDEDEHV-------------RAPIGHHQAGNCLMWACKACKRKSSTTDR 84
Query 61 RKVQSVGRLMRVFKTIREENELIMQLKGCT---PGDRIP 96
RK + MR + +++ N+ LK CT P R+P
Sbjct 85 RKAAT----MRERRRLKKVNQAFETLKRCTTTNPNQRLP 119
> hsa:54861 SNRK, DKFZp779A1866, FLJ20224, HSNFRK, KIAA0096; SNF
related kinase (EC:2.7.11.1); K08802 SNF related kinase [EC:2.7.11.1]
Length=765
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 18/40 (45%), Positives = 23/40 (57%), Gaps = 4/40 (10%)
Query 4 ILNPSLEEGEEDEEIDNVE-LPPEVVKIMRANIVLAEEGT 42
+LN EEGE D+E D E LPP K+ R + +A GT
Sbjct 485 VLNQIFEEGESDDEFDMDENLPP---KLSRLKMNIASPGT 521
> ath:AT3G59980 tRNA-binding region domain-containing protein
Length=273
Score = 28.9 bits (63), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 61 RKVQSVGRLMRVFKTIREENELIMQLKGCTPGDRIPVG 98
R V+S G L+ E EL++ +G PGDR+ G
Sbjct 173 RGVKSCGMLLAASDAAHENVELLVPPEGSVPGDRVWFG 210
> mmu:20623 Snrk, 2010012F07Rik, AI448042, AW547029, E030034B15,
MGC28970, R74830, mKIAA0096; SNF related kinase (EC:2.7.11.1);
K08802 SNF related kinase [EC:2.7.11.1]
Length=748
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 18/40 (45%), Positives = 23/40 (57%), Gaps = 4/40 (10%)
Query 4 ILNPSLEEGEEDEEIDNVE-LPPEVVKIMRANIVLAEEGT 42
+LN EEGE D+E D E LPP K+ R + +A GT
Sbjct 485 VLNQIFEEGESDDEFDMDENLPP---KLSRLKMNIASPGT 521
> bbo:BBOV_I005550 26.m00079; variant erythrocyte surface antigen-1
Length=1335
Score = 28.5 bits (62), Expect = 5.5, Method: Composition-based stats.
Identities = 25/97 (25%), Positives = 42/97 (43%), Gaps = 13/97 (13%)
Query 1 LYGILNPSLEEGEEDEEIDNVELPPEVVKIMRAN-------IVLAEEGTAPRSAGTPISR 53
LYGIL ++EG ED+ DN+ + + + N V+ + G P SA IS
Sbjct 758 LYGILEEKVDEGVEDKLKDNLTKVKDALNALTTNGGNGKLKDVVDKLGNDPSSAKDNISA 817
Query 54 ------ERAEALRRKVQSVGRLMRVFKTIREENELIM 84
E AL + +++ G+ K + E ++
Sbjct 818 AIHNVLEVLRALMKGLEAAGKNEEAKKKLGNHQEALL 854
Lambda K H
0.314 0.137 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2072286120
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40