bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5838_orf1
Length=140
Score E
Sequences producing significant alignments: (Bits) Value
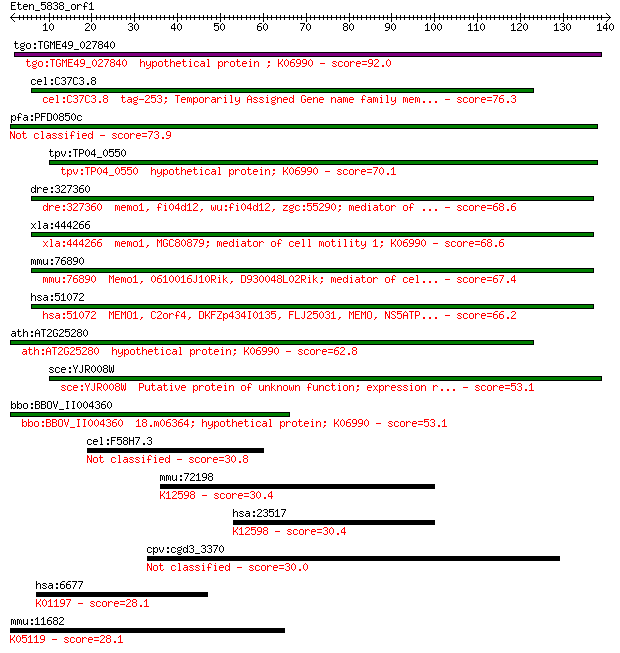
tgo:TGME49_027840 hypothetical protein ; K06990 92.0 6e-19
cel:C37C3.8 tag-253; Temporarily Assigned Gene name family mem... 76.3 3e-14
pfa:PFD0850c Memo-like protein 73.9 2e-13
tpv:TP04_0550 hypothetical protein; K06990 70.1 2e-12
dre:327360 memo1, fi04d12, wu:fi04d12, zgc:55290; mediator of ... 68.6 6e-12
xla:444266 memo1, MGC80879; mediator of cell motility 1; K06990 68.6 6e-12
mmu:76890 Memo1, 0610016J10Rik, D930048L02Rik; mediator of cel... 67.4 1e-11
hsa:51072 MEMO1, C2orf4, DKFZp434I0135, FLJ25031, MEMO, NS5ATP... 66.2 3e-11
ath:AT2G25280 hypothetical protein; K06990 62.8 3e-10
sce:YJR008W Putative protein of unknown function; expression r... 53.1 2e-07
bbo:BBOV_II004360 18.m06364; hypothetical protein; K06990 53.1 2e-07
cel:F58H7.3 lgc-30; Ligand-Gated ion Channel family member (lg... 30.8 1.4
mmu:72198 Skiv2l2, 2610528A15Rik, mKIAA0052; superkiller viral... 30.4 1.7
hsa:23517 SKIV2L2, Dob1, KIAA0052, MGC142069, Mtr4, fSAP118; s... 30.4 1.8
cpv:cgd3_3370 hypothetical protein 30.0 2.1
hsa:6677 SPAM1, HYA1, HYAL1, HYAL3, HYAL5, MGC26532, PH-20, PH... 28.1 8.7
mmu:11682 Alk, CD246, Tcrz; anaplastic lymphoma kinase (EC:2.7... 28.1 9.5
> tgo:TGME49_027840 hypothetical protein ; K06990
Length=302
Score = 92.0 bits (227), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 52/137 (37%), Positives = 79/137 (57%), Gaps = 6/137 (4%)
Query 2 GLSMYAETLVPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDKSLPLYKKIQALDREAI 61
G + A+ L P+F+ LFV S+D+ HWG R Y+YLP SLP++++I LD+E
Sbjct 161 GHAAVAKALRPYFLQEGNLFVFSSDFCHWGRRFRYSYLPP-ATASLPIFERIGILDKEGA 219
Query 62 DCVVNLNGSCLEEHVAKTGNRICGYDALLLFLRLLERVAESSGQRSEDLFSASLLSYSQS 121
+ + + +E+ +TGN ICG++ + +FL LL E+SG R F LL YSQS
Sbjct 220 ALIEQQDPAGFQEYYERTGNTICGHNPISIFLHLL----EASG-RPRSAFKTKLLDYSQS 274
Query 122 SLIRSASENAVGYVAIG 138
S + + S ++V Y A
Sbjct 275 SQVENESSSSVSYAAFA 291
> cel:C37C3.8 tag-253; Temporarily Assigned Gene name family member
(tag-253); K06990
Length=350
Score = 76.3 bits (186), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 38/117 (32%), Positives = 64/117 (54%), Gaps = 6/117 (5%)
Query 6 YAETLVPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDKSLPLYKKIQALDREAIDCVV 65
Y + DP LFVIS+D+ HWG R S++ P + S+P+Y++I +D++ + +
Sbjct 218 YGNIFAHYMEDPRNLFVISSDFCHWGERFSFS--PYDRHSSIPIYEQITNMDKQGMSAIE 275
Query 66 NLNGSCLEEHVAKTGNRICGYDALLLFLRLLERVAESSGQRSEDLFSASLLSYSQSS 122
LN + +++ KT N ICG + +L+ L+ E S+ E F L Y+QS+
Sbjct 276 TLNPAAFNDYLKKTQNTICGRNPILIMLQAAEHFRISNNHTHEFRF----LHYTQSN 328
> pfa:PFD0850c Memo-like protein
Length=296
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 36/137 (26%), Positives = 78/137 (56%), Gaps = 9/137 (6%)
Query 1 EGLSMYAETLVPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDKSLPLYKKIQALDREA 60
+ + ++A L +F D LF+ S+D+ H+GPR +T + + + ++K+I+ +D++A
Sbjct 164 KKIDLFANPLKKYFQDQHNLFLFSSDFCHYGPRFRFTNILQKYSDTF-IFKQIENMDKDA 222
Query 61 IDCVVNLNGSCLEEHVAKTGNRICGYDALLLFLRLLERVAESSGQRSEDLFSASLLSYSQ 120
++ + + + + +++ +T N ICG + + + L LL+ + S S L+ YSQ
Sbjct 223 VNIISHHDLTGFVDYLNETHNTICGSNPIKIMLNLLQHYSAS--------VSTKLMHYSQ 274
Query 121 SSLIRSASENAVGYVAI 137
S+ +S S+++V Y +
Sbjct 275 SNHAKSRSDSSVSYAGV 291
> tpv:TP04_0550 hypothetical protein; K06990
Length=290
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 47/131 (35%), Positives = 68/131 (51%), Gaps = 15/131 (11%)
Query 10 LVPFFMDPDTLFVISTDWSHWGPRSSYT---YLPENVDKSLPLYKKIQALDREAIDCVVN 66
LVP+F D TLFV S+D+ H+G R ++ Y EN PLY+KI+ LD+ ID +VN
Sbjct 166 LVPYFNDERTLFVFSSDFCHFGSRFQFSITGYESENK----PLYEKIEMLDKRGIDLIVN 221
Query 67 LNGSCLEEHVAKTGNRICGYDALLLFLRLLERVAESSGQRSEDLFSASLLSYSQSSLIRS 126
++ +T N ICG + + + S ++ LL YSQSS I
Sbjct 222 HKHDDFLWYLTETENTICGRNPI--------LLLLRLLAASNLNVTSRLLHYSQSSRITR 273
Query 127 ASENAVGYVAI 137
S+++V Y AI
Sbjct 274 VSDSSVSYAAI 284
> dre:327360 memo1, fi04d12, wu:fi04d12, zgc:55290; mediator of
cell motility 1; K06990
Length=297
Score = 68.6 bits (166), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 44/131 (33%), Positives = 68/131 (51%), Gaps = 8/131 (6%)
Query 6 YAETLVPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDKSLPLYKKIQALDREAIDCVV 65
Y + L + DP LF+IS D+ HWG R YTY E+ + +Y+ I+ LD+ + +
Sbjct 169 YGKLLSKYLADPSNLFIISPDFCHWGQRFRYTYYDESQGE---IYRSIEHLDKMGMGIIE 225
Query 66 NLNGSCLEEHVAKTGNRICGYDALLLFLRLLERVAESSGQRSEDLFSASLLSYSQSSLIR 125
L+ ++ K N ICG + + LL VAE + F S L+Y+QSS R
Sbjct 226 QLDPISFSNYLKKYHNTICGRHPIGV---LLNAVAELKKNGIDMNF--SFLNYAQSSQCR 280
Query 126 SASENAVGYVA 136
+ S+++V Y A
Sbjct 281 NWSDSSVSYAA 291
> xla:444266 memo1, MGC80879; mediator of cell motility 1; K06990
Length=297
Score = 68.6 bits (166), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 41/131 (31%), Positives = 67/131 (51%), Gaps = 8/131 (6%)
Query 6 YAETLVPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDKSLPLYKKIQALDREAIDCVV 65
+ + + DP LFVIS+D+ HWG R YTY E+ + +Y+ I+ LD+ + +
Sbjct 169 FGKVFSKYLADPTNLFVISSDFCHWGQRFRYTYYDESQGE---IYRSIENLDKMGMSIIE 225
Query 66 NLNGSCLEEHVAKTGNRICGYDALLLFLRLLERVAESSGQRSEDLFSASLLSYSQSSLIR 125
L+ ++ K N ICG + + L A + Q++ S S L+Y+QSS R
Sbjct 226 QLDPVQFSNYLKKYHNTICGRHPIGVLLN-----AATELQKNGVNMSFSFLNYAQSSQCR 280
Query 126 SASENAVGYVA 136
S +++V Y A
Sbjct 281 SWQDSSVSYAA 291
> mmu:76890 Memo1, 0610016J10Rik, D930048L02Rik; mediator of cell
motility 1; K06990
Length=297
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 38/131 (29%), Positives = 67/131 (51%), Gaps = 8/131 (6%)
Query 6 YAETLVPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDKSLPLYKKIQALDREAIDCVV 65
+ + + DP LFV+S+D+ HWG R Y+Y E+ + +Y+ I+ LD+ + +
Sbjct 169 FGKLFSKYLADPSNLFVVSSDFCHWGQRFRYSYYDESQGE---IYRSIEHLDKMGMSIIE 225
Query 66 NLNGSCLEEHVAKTGNRICGYDALLLFLRLLERVAESSGQRSEDLFSASLLSYSQSSLIR 125
L+ ++ K N ICG + + L + + Q++ S S L+Y+QSS R
Sbjct 226 QLDPVSFSNYLKKYHNTICGRHPIGVLLNAITEL-----QKNGMNMSFSFLNYAQSSQCR 280
Query 126 SASENAVGYVA 136
S +++V Y A
Sbjct 281 SWQDSSVSYAA 291
> hsa:51072 MEMO1, C2orf4, DKFZp434I0135, FLJ25031, MEMO, NS5ATP7;
mediator of cell motility 1; K06990
Length=274
Score = 66.2 bits (160), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 37/131 (28%), Positives = 67/131 (51%), Gaps = 8/131 (6%)
Query 6 YAETLVPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDKSLPLYKKIQALDREAIDCVV 65
+ + + DP LFV+S+D+ HWG R Y+Y E+ + +Y+ I+ LD+ + +
Sbjct 146 FGKLFSKYLADPSNLFVVSSDFCHWGQRFRYSYYDESQGE---IYRSIEHLDKMGMSIIE 202
Query 66 NLNGSCLEEHVAKTGNRICGYDALLLFLRLLERVAESSGQRSEDLFSASLLSYSQSSLIR 125
L+ ++ K N ICG + + L + + Q++ S S L+Y+QSS R
Sbjct 203 QLDPVSFSNYLKKYHNTICGRHPIGVLLNAITEL-----QKNGMNMSFSFLNYAQSSQCR 257
Query 126 SASENAVGYVA 136
+ +++V Y A
Sbjct 258 NWQDSSVSYAA 268
> ath:AT2G25280 hypothetical protein; K06990
Length=291
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 35/122 (28%), Positives = 62/122 (50%), Gaps = 11/122 (9%)
Query 1 EGLSMYAETLVPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDKSLPLYKKIQALDREA 60
E +MY E L + DP F +S+D+ HWG R +Y + +N + ++K I+ALD++
Sbjct 160 ENEAMYGELLAKYVDDPKNFFSVSSDFCHWGSRFNYMHY-DNTHGA--IHKSIEALDKKG 216
Query 61 IDCVVNLNGSCLEEHVAKTGNRICGYDALLLFLRLLERVAESSGQRSEDLFSASLLSYSQ 120
+D + + ++++ + N ICG + +FL +L + S + L Y Q
Sbjct 217 MDIIETGDPDAFKKYLLEFENTICGRHPISIFLHML--------KHSSSKIKINFLRYEQ 268
Query 121 SS 122
SS
Sbjct 269 SS 270
> sce:YJR008W Putative protein of unknown function; expression
repressed by inosine and choline in an Opi1p-dependent manner;
expression induced by mild heat-stress on a non-fermentable
carbon source; K06990
Length=338
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 44/162 (27%), Positives = 76/162 (46%), Gaps = 39/162 (24%)
Query 10 LVPFFMDPDTLFVISTDWSHWGPRSSYT-----------YLPENVD------------KS 46
L + DP+ LF++S+D+ HWG R YT + E +
Sbjct 183 LSEYIKDPNNLFIVSSDFCHWGRRFQYTGYVGSKEELNDAIQEETEVEMLTARSKLSHHQ 242
Query 47 LPLYKKIQALDREAIDCVVNL-NGS---CLEEHVAKTGNRICGYDALLLFLRLLERV--A 100
+P+++ I+ +DR A+ + + NG ++++ TGN ICG + + L L ++ A
Sbjct 243 VPIWQSIEIMDRYAMKTLSDTPNGERYDAWKQYLEITGNTICGEKPISVILSALSKIRDA 302
Query 101 ESSGQRSEDLFSASLLSYSQSSLIRSASENAV----GYVAIG 138
SG + + +YSQSS + S +++V GYV IG
Sbjct 303 GPSGIKFQ------WPNYSQSSHVTSIDDSSVSYASGYVTIG 338
> bbo:BBOV_II004360 18.m06364; hypothetical protein; K06990
Length=245
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 41/65 (63%), Gaps = 1/65 (1%)
Query 1 EGLSMYAETLVPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDKSLPLYKKIQALDREA 60
EGL + L+P+F DT+FVIS+D+ H+G R +T D+ +P++K I++LD +
Sbjct 137 EGLEDVGQALLPYFEKEDTIFVISSDFCHFGKRFGFTRTGFE-DQDMPIWKAIESLDLDG 195
Query 61 IDCVV 65
+ +V
Sbjct 196 VKLIV 200
> cel:F58H7.3 lgc-30; Ligand-Gated ion Channel family member (lgc-30)
Length=529
Score = 30.8 bits (68), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 14/46 (30%), Positives = 27/46 (58%), Gaps = 5/46 (10%)
Query 19 TLFVISTDWSHWGPRSSYTYLPENVDKSLP-----LYKKIQALDRE 59
++F ++T WS WG + S ++P+ + K P + + +ALD+E
Sbjct 5 SIFFMATIWSCWGAQESEGFIPKKLIKKPPAIDDTAFHRRRALDKE 50
> mmu:72198 Skiv2l2, 2610528A15Rik, mKIAA0052; superkiller viralicidic
activity 2-like 2 (S. cerevisiae) (EC:3.6.4.13); K12598
ATP-dependent RNA helicase DOB1 [EC:3.6.4.13]
Length=1040
Score = 30.4 bits (67), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 29/65 (44%), Gaps = 2/65 (3%)
Query 36 YTYLPENVDKSLPLYKKI-QALDREAIDCVVNLNGSCLEEHVAKTGNRICGYDALLLFLR 94
Y L V K+ Y I A REAI CV N N S L G +C A+ L LR
Sbjct 120 YIPLKPRVGKAAKEYPFILDAFQREAIQCVDN-NQSVLVSAHTSAGKTVCAEYAIALALR 178
Query 95 LLERV 99
+RV
Sbjct 179 EKQRV 183
> hsa:23517 SKIV2L2, Dob1, KIAA0052, MGC142069, Mtr4, fSAP118;
superkiller viralicidic activity 2-like 2 (S. cerevisiae) (EC:3.6.4.13);
K12598 ATP-dependent RNA helicase DOB1 [EC:3.6.4.13]
Length=1042
Score = 30.4 bits (67), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 23/47 (48%), Gaps = 1/47 (2%)
Query 53 IQALDREAIDCVVNLNGSCLEEHVAKTGNRICGYDALLLFLRLLERV 99
+ A REAI CV N N S L G +C A+ L LR +RV
Sbjct 140 LDAFQREAIQCVDN-NQSVLVSAHTSAGKTVCAEYAIALALREKQRV 185
> cpv:cgd3_3370 hypothetical protein
Length=1507
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 44/99 (44%), Gaps = 4/99 (4%)
Query 33 RSSYTY-LPENVDKSLPLYKKIQALDREAIDCVVNLNGSCLEEHVAKTGNRICGYDALLL 91
+S Y+Y L NV+ P+ K+ A C +++ G C V GN+ DA
Sbjct 934 KSPYSYSLSNNVNTWFPVSKEFYAPRSGDNSCTLSIYGVCTTSKVYMPGNKPVNNDA-KF 992
Query 92 FLRL--LERVAESSGQRSEDLFSASLLSYSQSSLIRSAS 128
F+R ++ S + ED + L +S +S + S S
Sbjct 993 FIRFYSFQKNTFSLSFKREDGTIVAQLEFSSTSTLWSGS 1031
> hsa:6677 SPAM1, HYA1, HYAL1, HYAL3, HYAL5, MGC26532, PH-20,
PH20, SPAG15; sperm adhesion molecule 1 (PH-20 hyaluronidase,
zona pellucida binding) (EC:3.2.1.35); K01197 hyaluronoglucosaminidase
[EC:3.2.1.35]
Length=509
Score = 28.1 bits (61), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 12/40 (30%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 7 AETLVPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDKS 46
A+ + F+M D L + DW W P + + P++V K+
Sbjct 127 AKKDITFYMPVDNLGMAVIDWEEWRPTWARNWKPKDVYKN 166
> mmu:11682 Alk, CD246, Tcrz; anaplastic lymphoma kinase (EC:2.7.10.1);
K05119 anaplastic lymphoma kinase [EC:2.7.10.1]
Length=1621
Score = 28.1 bits (61), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 18/64 (28%), Positives = 31/64 (48%), Gaps = 2/64 (3%)
Query 1 EGLSMYAETLVPFFMDPDTLFVISTDWSHWGPRSSYTYLPENVDKSLPLYKKIQALDREA 60
EGLS++ T++ D ++ W WGP S T +N+ SL Y I ++ +
Sbjct 592 EGLSLWQHTVLSLLDVTDRFWLQIVTW--WGPGSRATVGFDNISISLDCYLTISGEEKMS 649
Query 61 IDCV 64
++ V
Sbjct 650 LNSV 653
Lambda K H
0.319 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2552834388
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40