bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5888_orf1
Length=100
Score E
Sequences producing significant alignments: (Bits) Value
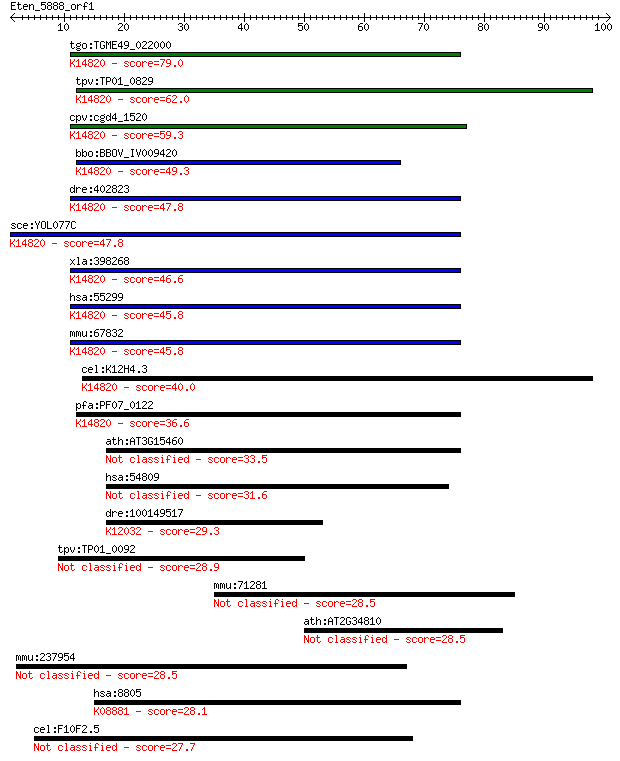
tgo:TGME49_022000 ribosome biogenesis protein Brix, putative ;... 79.0 4e-15
tpv:TP01_0829 hypothetical protein; K14820 ribosome biogenesis... 62.0 5e-10
cpv:cgd4_1520 Brx1p nucleolar protein required for biogenesis ... 59.3 3e-09
bbo:BBOV_IV009420 23.m05729; ribosome biogenesis Brix protein;... 49.3 3e-06
dre:402823 bxdc2; brix domain containing 2; K14820 ribosome bi... 47.8 8e-06
sce:YOL077C BRX1; Brx1p; K14820 ribosome biogenesis protein BRX1 47.8 8e-06
xla:398268 brix1, brix, bxdc2; BRX1, biogenesis of ribosomes, ... 46.6 2e-05
hsa:55299 BRIX1, BRIX, BXDC2, FLJ11100; BRX1, biogenesis of ri... 45.8 3e-05
mmu:67832 Brix1, 1110064N10Rik, Bxdc2, C76935; BRX1, biogenesi... 45.8 3e-05
cel:K12H4.3 hypothetical protein; K14820 ribosome biogenesis p... 40.0 0.002
pfa:PF07_0122 nucleolus BRIX protein, putative; K14820 ribosom... 36.6 0.022
ath:AT3G15460 brix domain-containing protein 33.5 0.18
hsa:54809 SAMD9, C7orf5, DRIF1, FLJ20073, KIAA2004, NFTC, OEF1... 31.6 0.57
dre:100149517 tripartite motif-containing 28 protein-like; K12... 29.3 3.3
tpv:TP01_0092 hypothetical protein 28.9 3.9
mmu:71281 Apobec4, 4933431M11Rik; apolipoprotein B mRNA editin... 28.5 5.0
ath:AT2G34810 FAD-binding domain-containing protein 28.5 5.4
mmu:237954 Lrrc37a, Gm251; leucine rich repeat containing 37A 28.5 5.6
hsa:8805 TRIM24, PTC6, RNF82, TF1A, TIF1, TIF1A, TIF1ALPHA, hT... 28.1 7.2
cel:F10F2.5 clec-154; C-type LECtin family member (clec-154) 27.7 8.4
> tgo:TGME49_022000 ribosome biogenesis protein Brix, putative
; K14820 ribosome biogenesis protein BRX1
Length=517
Score = 79.0 bits (193), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 34/67 (50%), Positives = 51/67 (76%), Gaps = 2/67 (2%)
Query 11 SQTKWEKKEKLQSIAELCQLRSCNSVLYLEQRKNDALLHVAKVPLGPTFIARLMN--GLE 68
+++KWE+K+ ++ I+EL QLR+CN+VLY E RK D LLHV+K+P GPT I R++N L
Sbjct 262 AESKWERKQTMKDISELAQLRACNNVLYFESRKRDLLLHVSKIPHGPTLIFRVLNIHTLA 321
Query 69 VLRFSGN 75
++ +GN
Sbjct 322 EMKMTGN 328
> tpv:TP01_0829 hypothetical protein; K14820 ribosome biogenesis
protein BRX1
Length=294
Score = 62.0 bits (149), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 32/89 (35%), Positives = 51/89 (57%), Gaps = 3/89 (3%)
Query 12 QTKWEKKEKLQSIAELCQLRSCNSVLYLEQRKNDALLHVAKVPLGPTFIARLMN--GLEV 69
++KW+ K + I ELC L +C LYLE RKN+ +L +AK P GPT +R+ N L+
Sbjct 80 ESKWDSKAGYKEINELCDLNNCTGALYLESRKNEMILWMAKTPNGPTLKSRVTNIHTLKD 139
Query 70 LRFSGNT-ERTPPVGKLSAALSASANIRI 97
+F+GN R+ P+ + S +++
Sbjct 140 SKFTGNCLARSRPLLTFDKSFSDEPYLKL 168
> cpv:cgd4_1520 Brx1p nucleolar protein required for biogenesis
of the 60S ribosomal subunit ; K14820 ribosome biogenesis
protein BRX1
Length=353
Score = 59.3 bits (142), Expect = 3e-09, Method: Composition-based stats.
Identities = 29/69 (42%), Positives = 47/69 (68%), Gaps = 3/69 (4%)
Query 11 SQTKWEKKEKLQSIAELCQLRSCNSVLYLEQRKNDAL-LHVAKVPLGPTFIARLMN--GL 67
S+ KWEKK+ I+E+ +LRSCN+++ LE R+++ L + VAK P GP+ +++N L
Sbjct 98 SEVKWEKKQPFSEISEMSELRSCNNIILLEARRHEELYMWVAKCPNGPSIKFQILNIHTL 157
Query 68 EVLRFSGNT 76
LR +GN+
Sbjct 158 GELRLAGNS 166
> bbo:BBOV_IV009420 23.m05729; ribosome biogenesis Brix protein;
K14820 ribosome biogenesis protein BRX1
Length=281
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 37/54 (68%), Gaps = 0/54 (0%)
Query 12 QTKWEKKEKLQSIAELCQLRSCNSVLYLEQRKNDALLHVAKVPLGPTFIARLMN 65
++K EK++ +++ L +L CNSV+++E RK + ++ +A P GP+F AR+ N
Sbjct 125 ESKLEKRDSRKALNTLAELSRCNSVIFIENRKCEVIMWIALTPYGPSFKARITN 178
> dre:402823 bxdc2; brix domain containing 2; K14820 ribosome
biogenesis protein BRX1
Length=383
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 21/68 (30%), Positives = 45/68 (66%), Gaps = 3/68 (4%)
Query 11 SQTKWEKKEKLQSIAELCQLRSCNSVLYLE-QRKNDALLHVAKVPLGPT--FIARLMNGL 67
+ TK ++K+KL + E+C++++CN ++ E ++K D + ++ VP GP+ F+ + ++ L
Sbjct 126 ADTKMDRKDKLFVVNEVCEIKNCNKCIFFEAKKKQDLYMWISNVPHGPSVKFLVQNVHTL 185
Query 68 EVLRFSGN 75
L+ +GN
Sbjct 186 AELKMTGN 193
> sce:YOL077C BRX1; Brx1p; K14820 ribosome biogenesis protein
BRX1
Length=291
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 28/82 (34%), Positives = 48/82 (58%), Gaps = 7/82 (8%)
Query 1 HFVWALGFVLSQT----KWEKKEKLQSIAELCQLRSCNSVLYLEQRKN-DALLHVAKVPL 55
H + L +L + K + K+ LQ + E+ +L +CN+VL+ E RK+ D L ++K P
Sbjct 47 HLIQDLSGLLPHSRKEPKLDTKKDLQQLNEIAELYNCNNVLFFEARKHQDLYLWLSKPPN 106
Query 56 GPT--FIARLMNGLEVLRFSGN 75
GPT F + ++ ++ L F+GN
Sbjct 107 GPTIKFYIQNLHTMDELNFTGN 128
> xla:398268 brix1, brix, bxdc2; BRX1, biogenesis of ribosomes,
homolog; K14820 ribosome biogenesis protein BRX1
Length=339
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 45/68 (66%), Gaps = 3/68 (4%)
Query 11 SQTKWEKKEKLQSIAELCQLRSCNSVLYLE-QRKNDALLHVAKVPLGPT--FIARLMNGL 67
++TK ++K+KL + E+C++++CN +Y E ++K D + ++ P GP+ F+ + ++ L
Sbjct 83 AETKMDRKDKLFVVNEVCEMKNCNKCIYFEAKKKQDLYMWLSNSPEGPSAKFLVQNIHTL 142
Query 68 EVLRFSGN 75
L+ SGN
Sbjct 143 AELKMSGN 150
> hsa:55299 BRIX1, BRIX, BXDC2, FLJ11100; BRX1, biogenesis of
ribosomes, homolog (S. cerevisiae); K14820 ribosome biogenesis
protein BRX1
Length=353
Score = 45.8 bits (107), Expect = 3e-05, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 44/68 (64%), Gaps = 3/68 (4%)
Query 11 SQTKWEKKEKLQSIAELCQLRSCNSVLYLE-QRKNDALLHVAKVPLGPT--FIARLMNGL 67
+ TK ++K+KL I E+C++++CN +Y E ++K D + ++ P GP+ F+ + ++ L
Sbjct 90 ADTKMDRKDKLFVINEVCEMKNCNKCIYFEAKKKQDLYMWLSNSPHGPSAKFLVQNIHTL 149
Query 68 EVLRFSGN 75
L+ +GN
Sbjct 150 AELKMTGN 157
> mmu:67832 Brix1, 1110064N10Rik, Bxdc2, C76935; BRX1, biogenesis
of ribosomes, homolog (S. cerevisiae); K14820 ribosome biogenesis
protein BRX1
Length=353
Score = 45.8 bits (107), Expect = 3e-05, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 44/68 (64%), Gaps = 3/68 (4%)
Query 11 SQTKWEKKEKLQSIAELCQLRSCNSVLYLE-QRKNDALLHVAKVPLGPT--FIARLMNGL 67
+ TK ++K+KL I E+C++++CN +Y E ++K D + ++ P GP+ F+ + ++ L
Sbjct 90 ADTKMDRKDKLFVINEVCEMKNCNKCIYFEAKKKQDLYMWLSNSPHGPSAKFLVQNIHTL 149
Query 68 EVLRFSGN 75
L+ +GN
Sbjct 150 AELKMTGN 157
> cel:K12H4.3 hypothetical protein; K14820 ribosome biogenesis
protein BRX1
Length=352
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/89 (25%), Positives = 48/89 (53%), Gaps = 4/89 (4%)
Query 13 TKWEKKEKLQSIAELCQLRSCNSVLYLEQRK-NDALLHVAKVPLGPT--FIARLMNGLEV 69
+K ++++ L + E+ ++++C V+Y E RK D L ++ V GP+ F+ ++ ++
Sbjct 95 SKLDQQKSLNVLNEIAEMKNCTKVMYFESRKRKDTYLWMSNVEKGPSIKFLVHNVHTMKE 154
Query 70 LRFSGNTER-TPPVGKLSAALSASANIRI 97
L+ SGN R + PV A +++
Sbjct 155 LKMSGNCLRASRPVLSFDDAFDKKPQLKL 183
> pfa:PF07_0122 nucleolus BRIX protein, putative; K14820 ribosome
biogenesis protein BRX1
Length=416
Score = 36.6 bits (83), Expect = 0.022, Method: Composition-based stats.
Identities = 17/66 (25%), Positives = 35/66 (53%), Gaps = 2/66 (3%)
Query 12 QTKWEKKEKLQSIAELCQLRSCNSVLYLEQRKNDALLHVAKVPLGPTFIARLMN--GLEV 69
+ KW+KK + ++ R+CN++++ + ++ L + K GP+ +++ L
Sbjct 195 ENKWDKKVPKSELNDISYSRNCNNIIFFDVKRKRYCLWICKSITGPSLYFEILDYIPLHS 254
Query 70 LRFSGN 75
L FSGN
Sbjct 255 LLFSGN 260
> ath:AT3G15460 brix domain-containing protein
Length=315
Score = 33.5 bits (75), Expect = 0.18, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 36/62 (58%), Gaps = 3/62 (4%)
Query 17 KKEKLQSIAELCQLRSCNSVLYLEQRKN-DALLHVAKVPLGPT--FIARLMNGLEVLRFS 73
K + ++ EL +L+ +S L+ E RK+ D + + K P GP+ F+ ++ +E L+ +
Sbjct 90 KSSRGATLNELIELKGSSSCLFFECRKHKDLYMWMVKSPGGPSVKFLVNAVHTMEELKLT 149
Query 74 GN 75
GN
Sbjct 150 GN 151
> hsa:54809 SAMD9, C7orf5, DRIF1, FLJ20073, KIAA2004, NFTC, OEF1,
OEF2; sterile alpha motif domain containing 9
Length=1589
Score = 31.6 bits (70), Expect = 0.57, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 32/57 (56%), Gaps = 4/57 (7%)
Query 17 KKEKLQSIAELCQLRSCNSVLYLEQRKNDALLHVAKVPLGPTFIARLMNGLEVLRFS 73
K+EK+Q + Q R+ N+ LY+E N+ + +P+ P F+ +L +G + + S
Sbjct 1519 KEEKVQELLLRLQGRAENNCLYIEYGINEKI----TIPITPAFLGQLRSGRSIEKVS 1571
> dre:100149517 tripartite motif-containing 28 protein-like; K12032
tripartite motif-containing protein 66
Length=1062
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 14/36 (38%), Positives = 22/36 (61%), Gaps = 1/36 (2%)
Query 17 KKEKLQSIAELCQLRSCNSVLYLEQRKNDALLHVAK 52
K+E L+ + E C L +C S+ +L K+ L+HV K
Sbjct 6 KQEPLELLCETCDLMAC-SICHLSAHKDHRLVHVGK 40
> tpv:TP01_0092 hypothetical protein
Length=2607
Score = 28.9 bits (63), Expect = 3.9, Method: Composition-based stats.
Identities = 15/41 (36%), Positives = 24/41 (58%), Gaps = 0/41 (0%)
Query 9 VLSQTKWEKKEKLQSIAELCQLRSCNSVLYLEQRKNDALLH 49
V+ QT E +E L +++EL + V+ LEQ ++ LLH
Sbjct 656 VVCQTNEEIRETLLTVSELACVYQVKKVIDLEQSDDNFLLH 696
> mmu:71281 Apobec4, 4933431M11Rik; apolipoprotein B mRNA editing
enzyme, catalytic polypeptide-like 4 (putative)
Length=374
Score = 28.5 bits (62), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 14/50 (28%), Positives = 24/50 (48%), Gaps = 0/50 (0%)
Query 35 SVLYLEQRKNDALLHVAKVPLGPTFIARLMNGLEVLRFSGNTERTPPVGK 84
+VL L ++ +HV P P + R +N L++ F + PP G+
Sbjct 283 AVLMLVSNRDLPPIHVGSTPQKPRTVVRHLNMLQLSSFKVKDVKKPPSGR 332
> ath:AT2G34810 FAD-binding domain-containing protein
Length=540
Score = 28.5 bits (62), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 18/33 (54%), Gaps = 0/33 (0%)
Query 50 VAKVPLGPTFIARLMNGLEVLRFSGNTERTPPV 82
A +P +F LMNG+ LRF+ + R P V
Sbjct 53 TAVIPTNSSFSTNLMNGVRNLRFASVSTRKPEV 85
> mmu:237954 Lrrc37a, Gm251; leucine rich repeat containing 37A
Length=1214
Score = 28.5 bits (62), Expect = 5.6, Method: Composition-based stats.
Identities = 19/68 (27%), Positives = 34/68 (50%), Gaps = 3/68 (4%)
Query 2 FVWALGFVLSQTKWEK--KEKLQSIAELCQLR-SCNSVLYLEQRKNDALLHVAKVPLGPT 58
+ WA +LS+ + + KE + + L L SCN + Y+E+R ++L + + LG
Sbjct 315 YRWAEKLILSENRLTELHKESFEGLLTLQVLDLSCNKIHYIERRTFESLPFLKYINLGCN 374
Query 59 FIARLMNG 66
+ L G
Sbjct 375 LLTELNFG 382
> hsa:8805 TRIM24, PTC6, RNF82, TF1A, TIF1, TIF1A, TIF1ALPHA,
hTIF1; tripartite motif containing 24; K08881 tripartite motif-containing
protein 24
Length=1016
Score = 28.1 bits (61), Expect = 7.2, Method: Composition-based stats.
Identities = 18/66 (27%), Positives = 32/66 (48%), Gaps = 6/66 (9%)
Query 15 WEKKEKLQSIAELCQLRSCNSVLYLEQRKN-----DALLHVAKVPLGPTFIARLMNGLEV 69
+ KKE+L+ E C +C LE +++ + KV + T I +LM +
Sbjct 225 FHKKEQLKLYCETCDKLTCRDCQLLEHKEHRYQFIEEAFQNQKVIID-TLITKLMEKTKY 283
Query 70 LRFSGN 75
++F+GN
Sbjct 284 IKFTGN 289
> cel:F10F2.5 clec-154; C-type LECtin family member (clec-154)
Length=525
Score = 27.7 bits (60), Expect = 8.4, Method: Composition-based stats.
Identities = 16/63 (25%), Positives = 30/63 (47%), Gaps = 4/63 (6%)
Query 5 ALGFVLSQTKWEKKEKLQSIAELCQLRSCNSVLYLEQRKNDALLHVAKVPLGPTFIARLM 64
A+ F L W K E +S C + +S + L +R+N+ L ++ P + +++
Sbjct 304 AVPFPLDNAIWRKGEPNES----CDAATWSSAVVLSRRENEGLEVMSDARYAPIYCEQIV 359
Query 65 NGL 67
GL
Sbjct 360 KGL 362
Lambda K H
0.322 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2041372988
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40