bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5955_orf1
Length=71
Score E
Sequences producing significant alignments: (Bits) Value
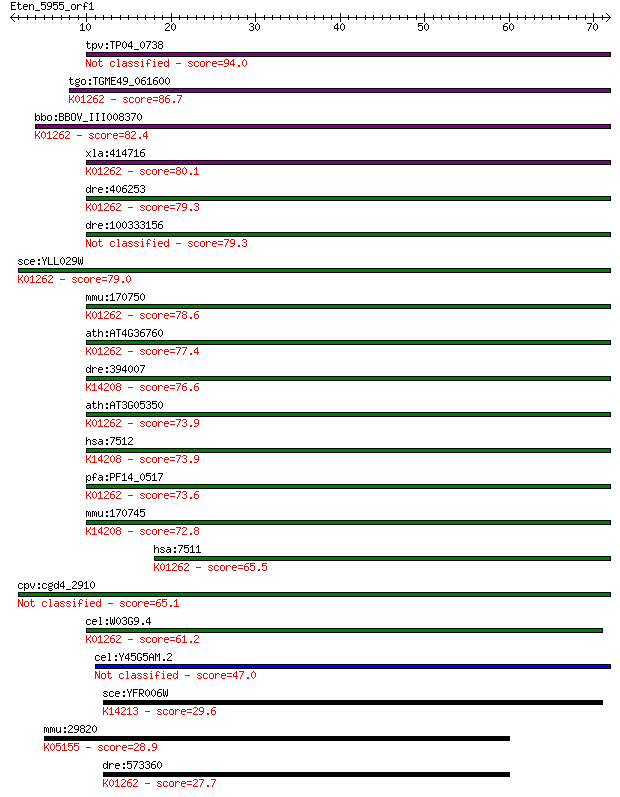
tpv:TP04_0738 peptidase 94.0 1e-19
tgo:TGME49_061600 X-prolyl aminopeptidase, putative (EC:3.4.11... 86.7 2e-17
bbo:BBOV_III008370 17.m07732; metallopeptidase M24 family prot... 82.4 3e-16
xla:414716 hypothetical protein MGC83093; K01262 Xaa-Pro amino... 80.1 1e-15
dre:406253 xpnpep1, MGC56366, fa02e02, wu:fa02e02, zgc:56366, ... 79.3 2e-15
dre:100333156 peptidase-like 79.3 3e-15
sce:YLL029W FRA1; Fra1p (EC:3.4.11.9); K01262 Xaa-Pro aminopep... 79.0 3e-15
mmu:170750 Xpnpep1, D230045I08Rik; X-prolyl aminopeptidase (am... 78.6 5e-15
ath:AT4G36760 ATAPP1; ATAPP1; N-1-naphthylphthalamic acid bind... 77.4 9e-15
dre:394007 xpnpep2, MGC63528, zgc:63528; X-prolyl aminopeptida... 76.6 2e-14
ath:AT3G05350 aminopeptidase/ hydrolase; K01262 Xaa-Pro aminop... 73.9 1e-13
hsa:7512 XPNPEP2, APP2; X-prolyl aminopeptidase (aminopeptidas... 73.9 1e-13
pfa:PF14_0517 peptidase, putative; K01262 Xaa-Pro aminopeptida... 73.6 2e-13
mmu:170745 Xpnpep2, 9030008G12Rik, mAPP; X-prolyl aminopeptida... 72.8 3e-13
hsa:7511 XPNPEP1, APP1, SAMP, XPNPEP, XPNPEPL, XPNPEPL1; X-pro... 65.5 4e-11
cpv:cgd4_2910 aminopeptidase 65.1 5e-11
cel:W03G9.4 app-1; AminoPeptidase P family member (app-1); K01... 61.2 7e-10
cel:Y45G5AM.2 hypothetical protein 47.0 1e-05
sce:YFR006W Putative X-Pro aminopeptidase; green fluorescent p... 29.6 2.2
mmu:29820 Tnfrsf19, AL023044, AW123854, TAJ, TAJ-ALPHA, TRADE,... 28.9 3.7
dre:573360 xpnpep3, MGC76905, zgc:76905; X-prolyl aminopeptida... 27.7 8.6
> tpv:TP04_0738 peptidase
Length=660
Score = 94.0 bits (232), Expect = 1e-19, Method: Composition-based stats.
Identities = 43/62 (69%), Positives = 50/62 (80%), Gaps = 0/62 (0%)
Query 10 VFLLDSGGQYLTGTTDVTRTLHLGEPTDEQKEAYTLVLKGMIGLSAQKFPEGSKGPQIDA 69
++LLDSGGQYLTGTTDVTRT+H G PTDE+K AYTLVLKG + L KFPEG+ G +D
Sbjct 441 MYLLDSGGQYLTGTTDVTRTVHFGTPTDEEKLAYTLVLKGHLALRHAKFPEGTPGESLDV 500
Query 70 LA 71
LA
Sbjct 501 LA 502
> tgo:TGME49_061600 X-prolyl aminopeptidase, putative (EC:3.4.11.9);
K01262 Xaa-Pro aminopeptidase [EC:3.4.11.9]
Length=724
Score = 86.7 bits (213), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 41/65 (63%), Positives = 51/65 (78%), Gaps = 1/65 (1%)
Query 8 SPVFLLDSGGQY-LTGTTDVTRTLHLGEPTDEQKEAYTLVLKGMIGLSAQKFPEGSKGPQ 66
S +FLLDSG Y + GTTDVTRT+H G P++ QK +TLVLKG IGLS Q FP+G++GPQ
Sbjct 486 SCLFLLDSGAHYAVGGTTDVTRTVHTGTPSESQKRYFTLVLKGFIGLSRQVFPQGTRGPQ 545
Query 67 IDALA 71
+D LA
Sbjct 546 LDVLA 550
> bbo:BBOV_III008370 17.m07732; metallopeptidase M24 family protein;
K01262 Xaa-Pro aminopeptidase [EC:3.4.11.9]
Length=624
Score = 82.4 bits (202), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 40/68 (58%), Positives = 49/68 (72%), Gaps = 0/68 (0%)
Query 4 AQNESPVFLLDSGGQYLTGTTDVTRTLHLGEPTDEQKEAYTLVLKGMIGLSAQKFPEGSK 63
A+ E ++LLDSGGQY GTTDVTRT+H G P+DE+KEAYT VLKG + L FPE +
Sbjct 399 AKIEPKIYLLDSGGQYPGGTTDVTRTIHFGTPSDEEKEAYTQVLKGHLALGHAIFPEHTS 458
Query 64 GPQIDALA 71
G +D LA
Sbjct 459 GATLDILA 466
> xla:414716 hypothetical protein MGC83093; K01262 Xaa-Pro aminopeptidase
[EC:3.4.11.9]
Length=621
Score = 80.1 bits (196), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 36/62 (58%), Positives = 46/62 (74%), Gaps = 0/62 (0%)
Query 10 VFLLDSGGQYLTGTTDVTRTLHLGEPTDEQKEAYTLVLKGMIGLSAQKFPEGSKGPQIDA 69
+FLLDSG Q+ GTTDVTRTLH G PTD +KE +T VL+G I +S+ FP G+KG +D+
Sbjct 409 IFLLDSGAQFKDGTTDVTRTLHFGTPTDYEKECFTYVLQGHIAVSSAVFPNGTKGHLLDS 468
Query 70 LA 71
A
Sbjct 469 FA 470
> dre:406253 xpnpep1, MGC56366, fa02e02, wu:fa02e02, zgc:56366,
zgc:77772; X-prolyl aminopeptidase (aminopeptidase P) 1, soluble
(EC:3.4.11.9); K01262 Xaa-Pro aminopeptidase [EC:3.4.11.9]
Length=620
Score = 79.3 bits (194), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 35/62 (56%), Positives = 46/62 (74%), Gaps = 0/62 (0%)
Query 10 VFLLDSGGQYLTGTTDVTRTLHLGEPTDEQKEAYTLVLKGMIGLSAQKFPEGSKGPQIDA 69
V+L+DSG QY GTTDVTRT+H G P++ +KE +T VLKG I +SA FP G+KG +D+
Sbjct 411 VYLIDSGAQYTDGTTDVTRTVHFGTPSEYEKECFTYVLKGHIAVSAAVFPNGTKGHLLDS 470
Query 70 LA 71
A
Sbjct 471 FA 472
> dre:100333156 peptidase-like
Length=611
Score = 79.3 bits (194), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 35/62 (56%), Positives = 48/62 (77%), Gaps = 0/62 (0%)
Query 10 VFLLDSGGQYLTGTTDVTRTLHLGEPTDEQKEAYTLVLKGMIGLSAQKFPEGSKGPQIDA 69
+FL+DSG QY+ GTTD+TRT+ +G EQK +TLVLKGMIG+S +FP+G++G +D
Sbjct 409 LFLIDSGAQYVNGTTDITRTVAIGAVPQEQKRFFTLVLKGMIGISTARFPKGTRGCDLDP 468
Query 70 LA 71
LA
Sbjct 469 LA 470
> sce:YLL029W FRA1; Fra1p (EC:3.4.11.9); K01262 Xaa-Pro aminopeptidase
[EC:3.4.11.9]
Length=749
Score = 79.0 bits (193), Expect = 3e-15, Method: Composition-based stats.
Identities = 38/75 (50%), Positives = 50/75 (66%), Gaps = 5/75 (6%)
Query 2 PPAQNES-----PVFLLDSGGQYLTGTTDVTRTLHLGEPTDEQKEAYTLVLKGMIGLSAQ 56
PP +N S ++L DSG Q+L GTTD+TRT+HL +PT E+ + YTLVLKG + L
Sbjct 534 PPVENSSMIDPTKIYLCDSGSQFLEGTTDITRTIHLTKPTKEEMDNYTLVLKGGLALERL 593
Query 57 KFPEGSKGPQIDALA 71
FPE + G IDA+A
Sbjct 594 IFPENTPGFNIDAIA 608
> mmu:170750 Xpnpep1, D230045I08Rik; X-prolyl aminopeptidase (aminopeptidase
P) 1, soluble (EC:3.4.11.9); K01262 Xaa-Pro aminopeptidase
[EC:3.4.11.9]
Length=623
Score = 78.6 bits (192), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 36/62 (58%), Positives = 45/62 (72%), Gaps = 0/62 (0%)
Query 10 VFLLDSGGQYLTGTTDVTRTLHLGEPTDEQKEAYTLVLKGMIGLSAQKFPEGSKGPQIDA 69
V+L+DSG QY GTTDVTRT+H G PT +KE +T VLKG I +SA FP G+KG +D+
Sbjct 411 VYLIDSGAQYKDGTTDVTRTMHFGTPTAYEKECFTYVLKGHIAVSAAVFPTGTKGHLLDS 470
Query 70 LA 71
A
Sbjct 471 FA 472
> ath:AT4G36760 ATAPP1; ATAPP1; N-1-naphthylphthalamic acid binding
/ aminopeptidase; K01262 Xaa-Pro aminopeptidase [EC:3.4.11.9]
Length=645
Score = 77.4 bits (189), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 32/62 (51%), Positives = 44/62 (70%), Gaps = 0/62 (0%)
Query 10 VFLLDSGGQYLTGTTDVTRTLHLGEPTDEQKEAYTLVLKGMIGLSAQKFPEGSKGPQIDA 69
++L DSG QYL GTTD+TRT+H G+P+ +KE YT V KG + L +FP+G+ G +D
Sbjct 436 IYLCDSGAQYLDGTTDITRTVHFGKPSAHEKECYTAVFKGHVALGNARFPKGTNGYTLDI 495
Query 70 LA 71
LA
Sbjct 496 LA 497
> dre:394007 xpnpep2, MGC63528, zgc:63528; X-prolyl aminopeptidase
(aminopeptidase P) 2, membrane-bound (EC:3.4.11.9); K14208
Xaa-Pro aminopeptidase 2 [EC:3.4.11.9]
Length=702
Score = 76.6 bits (187), Expect = 2e-14, Method: Composition-based stats.
Identities = 36/62 (58%), Positives = 47/62 (75%), Gaps = 0/62 (0%)
Query 10 VFLLDSGGQYLTGTTDVTRTLHLGEPTDEQKEAYTLVLKGMIGLSAQKFPEGSKGPQIDA 69
++L+DSGGQYL GTTD+TRT+H G+PTD QKEAYT VL G I +S FP G++G ++
Sbjct 472 MYLVDSGGQYLDGTTDITRTVHWGKPTDFQKEAYTRVLMGNIEISRTIFPAGTRGVYMEM 531
Query 70 LA 71
L
Sbjct 532 LG 533
> ath:AT3G05350 aminopeptidase/ hydrolase; K01262 Xaa-Pro aminopeptidase
[EC:3.4.11.9]
Length=710
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 33/62 (53%), Positives = 43/62 (69%), Gaps = 0/62 (0%)
Query 10 VFLLDSGGQYLTGTTDVTRTLHLGEPTDEQKEAYTLVLKGMIGLSAQKFPEGSKGPQIDA 69
+FLLDSG QY+ GTTD+TRT+H EP+ +KE +T VL+G I L FPEG+ G +D
Sbjct 502 LFLLDSGAQYVDGTTDITRTVHFSEPSAREKECFTRVLQGHIALDQAVFPEGTPGFVLDG 561
Query 70 LA 71
A
Sbjct 562 FA 563
> hsa:7512 XPNPEP2, APP2; X-prolyl aminopeptidase (aminopeptidase
P) 2, membrane-bound (EC:3.4.11.9); K14208 Xaa-Pro aminopeptidase
2 [EC:3.4.11.9]
Length=674
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 35/62 (56%), Positives = 43/62 (69%), Gaps = 0/62 (0%)
Query 10 VFLLDSGGQYLTGTTDVTRTLHLGEPTDEQKEAYTLVLKGMIGLSAQKFPEGSKGPQIDA 69
++LLDSGGQY GTTD+TRT+H G P+ QKEAYT VL G I LS FP + G ++A
Sbjct 446 MYLLDSGGQYWDGTTDITRTVHWGTPSAFQKEAYTRVLIGNIDLSRLIFPAATSGRMVEA 505
Query 70 LA 71
A
Sbjct 506 FA 507
> pfa:PF14_0517 peptidase, putative; K01262 Xaa-Pro aminopeptidase
[EC:3.4.11.9]
Length=764
Score = 73.6 bits (179), Expect = 2e-13, Method: Composition-based stats.
Identities = 35/62 (56%), Positives = 42/62 (67%), Gaps = 0/62 (0%)
Query 10 VFLLDSGGQYLTGTTDVTRTLHLGEPTDEQKEAYTLVLKGMIGLSAQKFPEGSKGPQIDA 69
++LLDSGGQYL GTTDVTRT H GEPT E+K YTLVLKG + L F + +D
Sbjct 553 IYLLDSGGQYLHGTTDVTRTTHFGEPTAEEKRIYTLVLKGHLRLRKVIFASYTNSSALDF 612
Query 70 LA 71
+A
Sbjct 613 IA 614
> mmu:170745 Xpnpep2, 9030008G12Rik, mAPP; X-prolyl aminopeptidase
(aminopeptidase P) 2, membrane-bound (EC:3.4.11.9); K14208
Xaa-Pro aminopeptidase 2 [EC:3.4.11.9]
Length=674
Score = 72.8 bits (177), Expect = 3e-13, Method: Composition-based stats.
Identities = 36/62 (58%), Positives = 43/62 (69%), Gaps = 0/62 (0%)
Query 10 VFLLDSGGQYLTGTTDVTRTLHLGEPTDEQKEAYTLVLKGMIGLSAQKFPEGSKGPQIDA 69
++L+DSGGQY GTTD+TRT+H G PT QKEAYT VL G I LS FP + G I+A
Sbjct 446 MYLVDSGGQYWDGTTDITRTVHWGTPTAFQKEAYTRVLMGNIDLSRLVFPAATSGRVIEA 505
Query 70 LA 71
A
Sbjct 506 FA 507
> hsa:7511 XPNPEP1, APP1, SAMP, XPNPEP, XPNPEPL, XPNPEPL1; X-prolyl
aminopeptidase (aminopeptidase P) 1, soluble (EC:3.4.11.9);
K01262 Xaa-Pro aminopeptidase [EC:3.4.11.9]
Length=642
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 30/54 (55%), Positives = 37/54 (68%), Gaps = 0/54 (0%)
Query 18 QYLTGTTDVTRTLHLGEPTDEQKEAYTLVLKGMIGLSAQKFPEGSKGPQIDALA 71
Y GTTDVTRT+H G PT +KE +T VLKG I +SA FP G+KG +D+ A
Sbjct 438 HYADGTTDVTRTMHFGTPTAYEKECFTYVLKGHIAVSAAVFPTGTKGHLLDSFA 491
> cpv:cgd4_2910 aminopeptidase
Length=694
Score = 65.1 bits (157), Expect = 5e-11, Method: Composition-based stats.
Identities = 41/79 (51%), Positives = 47/79 (59%), Gaps = 9/79 (11%)
Query 2 PPAQNESPV----FLLDSGGQYLTGTTDVTRTLHL---GE--PTDEQKEAYTLVLKGMIG 52
P +N S + +L DSGGQY TGTTDVTRTL L GE PT EQ E++T VL G I
Sbjct 451 PEKENSSIIKPDLYLCDSGGQYHTGTTDVTRTLFLFGIGEERPTIEQIESFTRVLIGFIR 510
Query 53 LSAQKFPEGSKGPQIDALA 71
L FP G+ ID LA
Sbjct 511 LHKLVFPIGTNATAIDVLA 529
> cel:W03G9.4 app-1; AminoPeptidase P family member (app-1); K01262
Xaa-Pro aminopeptidase [EC:3.4.11.9]
Length=616
Score = 61.2 bits (147), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 33/61 (54%), Positives = 38/61 (62%), Gaps = 0/61 (0%)
Query 10 VFLLDSGGQYLTGTTDVTRTLHLGEPTDEQKEAYTLVLKGMIGLSAQKFPEGSKGPQIDA 69
VFLLDSG Y GTTDVTRT+ P E TLVLKG I L+ KFP+G G ++D
Sbjct 409 VFLLDSGAHYGDGTTDVTRTVWYTNPPKEFILHNTLVLKGHINLARAKFPDGIYGSRLDT 468
Query 70 L 70
L
Sbjct 469 L 469
> cel:Y45G5AM.2 hypothetical protein
Length=1061
Score = 47.0 bits (110), Expect = 1e-05, Method: Composition-based stats.
Identities = 24/62 (38%), Positives = 35/62 (56%), Gaps = 1/62 (1%)
Query 11 FLLDSGGQYLTGTTDVTRTLHLGEPTDEQKEAYTLVLKGMIGLSAQKFPEG-SKGPQIDA 69
F+ +G Y G T+ RT+ PT+E YTLVLKG I L++ FP+ + G ++D
Sbjct 854 FMFQTGSHYTDGATNCARTIWDSYPTEEFMNQYTLVLKGHIRLASASFPKTLTYGSRLDI 913
Query 70 LA 71
A
Sbjct 914 FA 915
> sce:YFR006W Putative X-Pro aminopeptidase; green fluorescent
protein (GFP)-fusion protein localizes to the cytoplasm; YFR006W
is not an essential gene (EC:3.4.-.-); K14213 Xaa-Pro
dipeptidase [EC:3.4.13.9]
Length=535
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 19/60 (31%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query 12 LLDSGGQYLTGTTDVTRTLHL-GEPTDEQKEAYTLVLKGMIGLSAQKFPEGSKGPQIDAL 70
L+D+G ++ T+D+TR G+ T E +E Y VL M + ++ G+K + AL
Sbjct 314 LIDAGAEWRQYTSDITRCFPTSGKFTAEHREVYETVLD-MQNQAMERIKPGAKWDDLHAL 372
> mmu:29820 Tnfrsf19, AL023044, AW123854, TAJ, TAJ-ALPHA, TRADE,
Troy; tumor necrosis factor receptor superfamily, member
19; K05155 tumor necrosis factor receptor superfamily member
19
Length=416
Score = 28.9 bits (63), Expect = 3.7, Method: Composition-based stats.
Identities = 19/56 (33%), Positives = 28/56 (50%), Gaps = 1/56 (1%)
Query 5 QNESPVFLLDSGGQYLTGTTDVTRTLHLGEPTDEQKEAYT-LVLKGMIGLSAQKFP 59
+NES L SGGQ L GT + + ++ E TD + T V + + AQ+ P
Sbjct 339 ENESAASLDSSGGQDLAGTAALESSGNVSESTDSPRHGDTGTVWEQTLAQDAQRTP 394
> dre:573360 xpnpep3, MGC76905, zgc:76905; X-prolyl aminopeptidase
(aminopeptidase P) 3, putative (EC:3.4.11.9); K01262 Xaa-Pro
aminopeptidase [EC:3.4.11.9]
Length=510
Score = 27.7 bits (60), Expect = 8.6, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 27/49 (55%), Gaps = 1/49 (2%)
Query 12 LLDSGGQYLTGTTDVTRTLHL-GEPTDEQKEAYTLVLKGMIGLSAQKFP 59
LLD G +Y +D+TRT + G+ + Q+E Y VL+ + +Q P
Sbjct 329 LLDGGCEYFGYVSDITRTWPVNGKFSAAQRELYEAVLEVQLACLSQCSP 377
Lambda K H
0.310 0.133 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2020506228
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40