bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6057_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
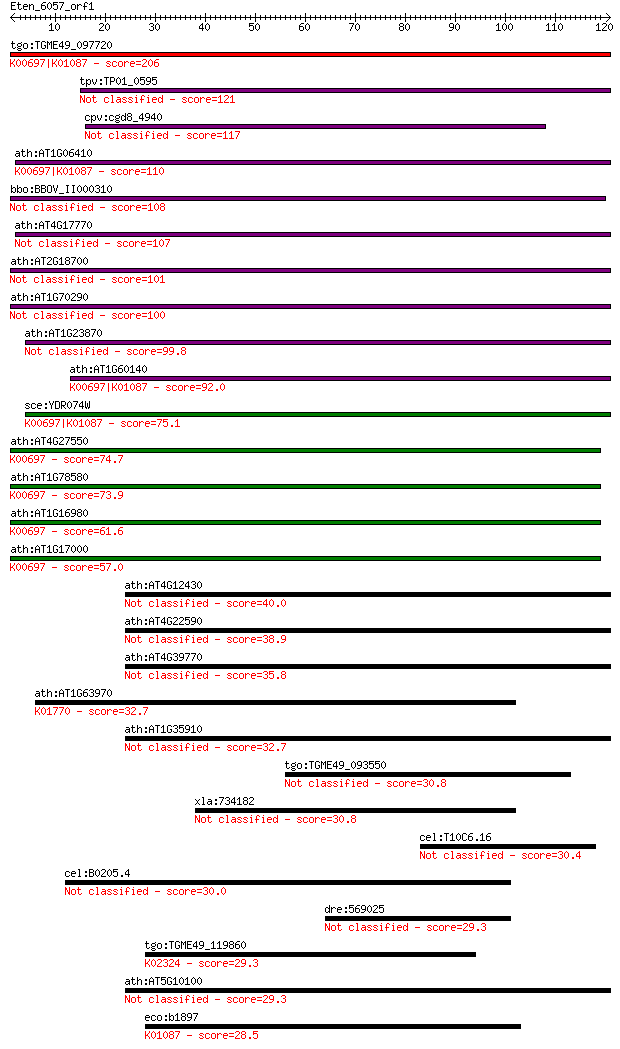
tgo:TGME49_097720 trehalose-6-phosphate synthase domain-contai... 206 2e-53
tpv:TP01_0595 trehalose-6-phosphate synthase 121 5e-28
cpv:cgd8_4940 trehalose-6-phosphate synthase of likely plant o... 117 8e-27
ath:AT1G06410 ATTPS7; ATTPS7; alpha,alpha-trehalose-phosphate ... 110 9e-25
bbo:BBOV_II000310 18.m06009; trehalose-6-phosphate synthase do... 108 5e-24
ath:AT4G17770 ATTPS5; ATTPS5; protein binding / transferase, t... 107 1e-23
ath:AT2G18700 ATTPS11; transferase, transferring glycosyl groups 101 6e-22
ath:AT1G70290 ATTPS8; ATTPS8; alpha,alpha-trehalose-phosphate ... 100 2e-21
ath:AT1G23870 ATTPS9; ATTPS9; transferase, transferring glycos... 99.8 2e-21
ath:AT1G60140 ATTPS10 (trehalose phosphate synthase); transfer... 92.0 4e-19
sce:YDR074W TPS2, HOG2, PFK3; Phosphatase subunit of the treha... 75.1 5e-14
ath:AT4G27550 ATTPS4; ATTPS4; alpha,alpha-trehalose-phosphate ... 74.7 7e-14
ath:AT1G78580 ATTPS1; ATTPS1 (TREHALOSE-6-PHOSPHATE SYNTHASE);... 73.9 1e-13
ath:AT1G16980 ATTPS2; ATTPS2; alpha,alpha-trehalose-phosphate ... 61.6 6e-10
ath:AT1G17000 ATTPS3; ATTPS3; alpha,alpha-trehalose-phosphate ... 57.0 1e-08
ath:AT4G12430 trehalose-6-phosphate phosphatase, putative (EC:... 40.0 0.002
ath:AT4G22590 trehalose-6-phosphate phosphatase, putative (EC:... 38.9 0.003
ath:AT4G39770 trehalose-6-phosphate phosphatase, putative (EC:... 35.8 0.032
ath:AT1G63970 ISPF; ISPF; 2-C-methyl-D-erythritol 2,4-cyclodip... 32.7 0.26
ath:AT1G35910 trehalose-6-phosphate phosphatase, putative 32.7 0.30
tgo:TGME49_093550 hypothetical protein 30.8 0.98
xla:734182 ara-55; Hic-5 30.8 1.1
cel:T10C6.16 hypothetical protein 30.4 1.6
cel:B0205.4 hypothetical protein 30.0 1.9
dre:569025 Sb:cb37 protein-like 29.3 3.2
tgo:TGME49_119860 DNA polymerase epsilon catalytic subunit, pu... 29.3 3.4
ath:AT5G10100 trehalose-6-phosphate phosphatase, putative (EC:... 29.3 3.5
eco:b1897 otsB, ECK1896, JW1886, otsP; trehalose-6-phosphate p... 28.5 5.6
> tgo:TGME49_097720 trehalose-6-phosphate synthase domain-containing
protein (EC:2.4.1.15 2.7.9.5 3.1.3.12); K00697 alpha,alpha-trehalose-phosphate
synthase (UDP-forming) [EC:2.4.1.15];
K01087 trehalose-phosphatase [EC:3.1.3.12]
Length=1222
Score = 206 bits (524), Expect = 2e-53, Method: Compositional matrix adjust.
Identities = 94/120 (78%), Positives = 106/120 (88%), Gaps = 0/120 (0%)
Query 1 TGDEWGCMCPDADFAWKSVAMELMLQYVKRTQGSFIENKGSALVFQYRDADPDFGQLQAK 60
TGD+W CM DF WK VA+ELMLQYVKRTQGSFIENKGSALVFQYRDADPDFG +QAK
Sbjct 863 TGDQWHCMSRQTDFTWKQVAIELMLQYVKRTQGSFIENKGSALVFQYRDADPDFGSMQAK 922
Query 61 DLSSHLSELLFGYPVTVVSGKGYVEVKLLGVNKGRAVERILRTLSTLHGDIDFIFCIGDD 120
DLS++L ELLFGYPV+V+SGKGYVEVKL GVNKG AVE++LR LS LHGD+DF+ C+GDD
Sbjct 923 DLSNYLGELLFGYPVSVMSGKGYVEVKLRGVNKGHAVEKVLRKLSNLHGDVDFVLCVGDD 982
> tpv:TP01_0595 trehalose-6-phosphate synthase
Length=1059
Score = 121 bits (304), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 54/106 (50%), Positives = 81/106 (76%), Gaps = 0/106 (0%)
Query 15 AWKSVAMELMLQYVKRTQGSFIENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYP 74
+W+ VA++L QYV+RT GS++E+K S LVFQ++++D +FG LQA +L+S L EL+ G+P
Sbjct 878 SWRPVALKLFEQYVQRTPGSYLESKDSCLVFQFQNSDQEFGILQANELNSSLCELMGGFP 937
Query 75 VTVVSGKGYVEVKLLGVNKGRAVERILRTLSTLHGDIDFIFCIGDD 120
V V GKGY+E++L G+NKG A+ ++ S+L GD DF+ C+GDD
Sbjct 938 VDVHRGKGYLELRLKGINKGNALVNVVHKYSSLFGDFDFVLCLGDD 983
> cpv:cgd8_4940 trehalose-6-phosphate synthase of likely plant
origin
Length=1417
Score = 117 bits (293), Expect = 8e-27, Method: Composition-based stats.
Identities = 53/92 (57%), Positives = 73/92 (79%), Gaps = 0/92 (0%)
Query 16 WKSVAMELMLQYVKRTQGSFIENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYPV 75
WK++ +LM QYV RTQGS+IENKG+ALVFQ++ +P FG QAK+LS++LSELL +PV
Sbjct 1212 WKNITFQLMEQYVLRTQGSYIENKGTALVFQFKYCEPYFGAWQAKELSNYLSELLINFPV 1271
Query 76 TVVSGKGYVEVKLLGVNKGRAVERILRTLSTL 107
V+SG +VEV+L G+NKG AV+ IL +++L
Sbjct 1272 NVISGNDFVEVRLQGINKGVAVQAILNQINSL 1303
> ath:AT1G06410 ATTPS7; ATTPS7; alpha,alpha-trehalose-phosphate
synthase (UDP-forming)/ transferase, transferring glycosyl
groups / trehalose-phosphatase; K00697 alpha,alpha-trehalose-phosphate
synthase (UDP-forming) [EC:2.4.1.15]; K01087 trehalose-phosphatase
[EC:3.1.3.12]
Length=851
Score = 110 bits (276), Expect = 9e-25, Method: Compositional matrix adjust.
Identities = 56/119 (47%), Positives = 72/119 (60%), Gaps = 0/119 (0%)
Query 2 GDEWGCMCPDADFAWKSVAMELMLQYVKRTQGSFIENKGSALVFQYRDADPDFGQLQAKD 61
+EW +DF W + +M QY + T GS IE K SALV+QYRDADP FG LQAK+
Sbjct 661 SEEWETCGQSSDFGWMQIVEPVMKQYTESTDGSSIEIKESALVWQYRDADPGFGSLQAKE 720
Query 62 LSSHLSELLFGYPVTVVSGKGYVEVKLLGVNKGRAVERILRTLSTLHGDIDFIFCIGDD 120
+ HL +L PV V SG VEVK GV+KG E+I +++ +DF+ CIGDD
Sbjct 721 MLEHLESVLANEPVAVKSGHYIVEVKPQGVSKGSVSEKIFSSMAGKGKPVDFVLCIGDD 779
> bbo:BBOV_II000310 18.m06009; trehalose-6-phosphate synthase
domain containing protein
Length=958
Score = 108 bits (269), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 51/119 (42%), Positives = 71/119 (59%), Gaps = 0/119 (0%)
Query 1 TGDEWGCMCPDADFAWKSVAMELMLQYVKRTQGSFIENKGSALVFQYRDADPDFGQLQAK 60
TG +W CM + D WK +A+++M QY RT GS+IEN +VFQY +DP+F Q+
Sbjct 770 TGGKWNCMVENVDDRWKVIALQIMEQYAHRTPGSYIENMDVMVVFQYHHSDPEFSATQSV 829
Query 61 DLSSHLSELLFGYPVTVVSGKGYVEVKLLGVNKGRAVERILRTLSTLHGDIDFIFCIGD 119
+L + L +++ YPV V K V V L GVNKG + I ++GD DFI C+GD
Sbjct 830 ELLTVLKQVMAPYPVDVQRTKWNVHVCLRGVNKGATLLNIAENYCRIYGDFDFILCVGD 888
> ath:AT4G17770 ATTPS5; ATTPS5; protein binding / transferase,
transferring glycosyl groups / trehalose-phosphatase
Length=862
Score = 107 bits (266), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 55/119 (46%), Positives = 72/119 (60%), Gaps = 0/119 (0%)
Query 2 GDEWGCMCPDADFAWKSVAMELMLQYVKRTQGSFIENKGSALVFQYRDADPDFGQLQAKD 61
G +W + F WK +A +M Y + T GS IE K +ALV+ Y+ ADPDFG QAK+
Sbjct 667 GTDWETSSLVSGFEWKQIAEPVMRLYTETTDGSTIETKETALVWNYQFADPDFGSCQAKE 726
Query 62 LSSHLSELLFGYPVTVVSGKGYVEVKLLGVNKGRAVERILRTLSTLHGDIDFIFCIGDD 120
L HL +L PV+V +G+ VEVK GVNKG ER+L T+ +DFI C+GDD
Sbjct 727 LMEHLESVLTNDPVSVKTGQQLVEVKPQGVNKGLVAERLLTTMQEKGKLLDFILCVGDD 785
> ath:AT2G18700 ATTPS11; transferase, transferring glycosyl groups
Length=862
Score = 101 bits (251), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 52/121 (42%), Positives = 72/121 (59%), Gaps = 1/121 (0%)
Query 1 TGDEWGCMCPDADFAWKSVAMELMLQYVKRTQGSFIENKGSALVFQYRDADPDFGQLQAK 60
+ W AD +WK +A +M Y++ T GSFIE K SA+V+ +++AD FG QAK
Sbjct 658 SNSPWETSELPADLSWKKIAKPVMNHYMEATDGSFIEEKESAMVWHHQEADHSFGSWQAK 717
Query 61 DLSSHLSELLFGYPVTVVSGKGYVEVKLLGVNKGRAVERILRTLSTLHGD-IDFIFCIGD 119
+L HL +L PV V G+ VEVK GV+KG+ VE ++ T+ G DF+ CIGD
Sbjct 718 ELLDHLESVLTNEPVVVKRGQHIVEVKPQGVSKGKVVEHLIATMRNTKGKRPDFLLCIGD 777
Query 120 D 120
D
Sbjct 778 D 778
> ath:AT1G70290 ATTPS8; ATTPS8; alpha,alpha-trehalose-phosphate
synthase (UDP-forming)/ transferase, transferring glycosyl
groups / trehalose-phosphatase
Length=856
Score = 100 bits (248), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 50/120 (41%), Positives = 71/120 (59%), Gaps = 0/120 (0%)
Query 1 TGDEWGCMCPDADFAWKSVAMELMLQYVKRTQGSFIENKGSALVFQYRDADPDFGQLQAK 60
+ DEW D W+S+ +M Y++ T G+ IE K SALV+ ++DADPDFG QAK
Sbjct 661 SKDEWETCYSPTDTEWRSMVEPVMRSYMEATDGTSIEFKESALVWHHQDADPDFGSCQAK 720
Query 61 DLSSHLSELLFGYPVTVVSGKGYVEVKLLGVNKGRAVERILRTLSTLHGDIDFIFCIGDD 120
++ HL +L PV V G+ VEVK GV+KG A E+++R + + + CIGDD
Sbjct 721 EMLDHLESVLANEPVVVKRGQHIVEVKPQGVSKGLAAEKVIREMVERGEPPEMVMCIGDD 780
> ath:AT1G23870 ATTPS9; ATTPS9; transferase, transferring glycosyl
groups / trehalose-phosphatase
Length=867
Score = 99.8 bits (247), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 52/117 (44%), Positives = 69/117 (58%), Gaps = 0/117 (0%)
Query 4 EWGCMCPDADFAWKSVAMELMLQYVKRTQGSFIENKGSALVFQYRDADPDFGQLQAKDLS 63
EW A+ WK++ +M Y+ T GS IE K SALV+ ++DADPDFG QAK+L
Sbjct 669 EWETCYSSAEAEWKTMVEPVMRSYMDATDGSTIEYKESALVWHHQDADPDFGACQAKELL 728
Query 64 SHLSELLFGYPVTVVSGKGYVEVKLLGVNKGRAVERILRTLSTLHGDIDFIFCIGDD 120
HL +L PV V G+ VEVK GV+KG AVE+++ + D + CIGDD
Sbjct 729 DHLESVLANEPVVVKRGQHIVEVKPQGVSKGLAVEKVIHQMVEDGNPPDMVMCIGDD 785
> ath:AT1G60140 ATTPS10 (trehalose phosphate synthase); transferase,
transferring glycosyl groups / trehalose-phosphatase;
K00697 alpha,alpha-trehalose-phosphate synthase (UDP-forming)
[EC:2.4.1.15]; K01087 trehalose-phosphatase [EC:3.1.3.12]
Length=861
Score = 92.0 bits (227), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 51/108 (47%), Positives = 62/108 (57%), Gaps = 0/108 (0%)
Query 13 DFAWKSVAMELMLQYVKRTQGSFIENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFG 72
D WK V +M Y + T GS IE K SALV+ ++DADPDFG QAK+L HL +L
Sbjct 678 DLEWKKVVEPIMRLYTETTDGSNIEAKESALVWHHQDADPDFGSCQAKELLDHLETVLVN 737
Query 73 YPVTVVSGKGYVEVKLLGVNKGRAVERILRTLSTLHGDIDFIFCIGDD 120
PV V G VEVK GV+KG +IL + DF+ CIGDD
Sbjct 738 EPVIVNRGHQIVEVKPQGVSKGLVTGKILSRMLEDGIAPDFVVCIGDD 785
> sce:YDR074W TPS2, HOG2, PFK3; Phosphatase subunit of the trehalose-6-phosphate
synthase/phosphatase complex, which synthesizes
the storage carbohydrate trehalose; expression is induced
by stress conditions and repressed by the Ras-cAMP pathway
(EC:3.1.3.12); K00697 alpha,alpha-trehalose-phosphate synthase
(UDP-forming) [EC:2.4.1.15]; K01087 trehalose-phosphatase
[EC:3.1.3.12]
Length=896
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 41/131 (31%), Positives = 63/131 (48%), Gaps = 14/131 (10%)
Query 4 EWGCMCPDADFAWKSVAMELMLQYVKRTQGSFIENKGSALVFQYRDADPDFGQLQAKDLS 63
+W + D +W+ E+M ++ RT GSFIE K AL + YR P+ G+ AK+L
Sbjct 652 DWVNLTEKVDMSWQVRVNEVMEEFTTRTPGSFIERKKVALTWHYRRTVPELGEFHAKELK 711
Query 64 SHLSELLFGYPVTVVSGKGYVEVKLLGVNKGRAVERILRTLSTLHGDI------------ 111
L + + V+ GK +EV+ VNKG V+R++ D+
Sbjct 712 EKLLSFTDDFDLEVMDGKANIEVRPRFVNKGEIVKRLVWHQHGKPQDMLKGISEKLPKDE 771
Query 112 --DFIFCIGDD 120
DF+ C+GDD
Sbjct 772 MPDFVLCLGDD 782
> ath:AT4G27550 ATTPS4; ATTPS4; alpha,alpha-trehalose-phosphate
synthase (UDP-forming)/ transferase, transferring glycosyl
groups; K00697 alpha,alpha-trehalose-phosphate synthase (UDP-forming)
[EC:2.4.1.15]
Length=795
Score = 74.7 bits (182), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 43/123 (34%), Positives = 67/123 (54%), Gaps = 5/123 (4%)
Query 1 TGDEWGCMCPD-ADFAWKSVAMELMLQYVKRTQGSFIENKGSALVFQYRDADPDFGQLQA 59
T EW P+ + W + + +RT GS++E ++LV+ Y +AD +FG+ QA
Sbjct 591 TSGEWVTRIPEHMNLEWIDGVKHVFKYFTERTPGSYLETSEASLVWNYENADAEFGRAQA 650
Query 60 KDLSSHL-SELLFGYPVTVVSGKGYVEVKLLGVNKGRAVERILRTL---STLHGDIDFIF 115
+D+ HL + + V VV G VEV +GV KG A+ERIL + ++ ID++
Sbjct 651 RDMLQHLWAGPISNASVDVVRGGQSVEVHAVGVTKGSAMERILGEIVHNKSMATPIDYVL 710
Query 116 CIG 118
CIG
Sbjct 711 CIG 713
> ath:AT1G78580 ATTPS1; ATTPS1 (TREHALOSE-6-PHOSPHATE SYNTHASE);
alpha,alpha-trehalose-phosphate synthase (UDP-forming)/ transferase,
transferring glycosyl groups; K00697 alpha,alpha-trehalose-phosphate
synthase (UDP-forming) [EC:2.4.1.15]
Length=942
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 42/123 (34%), Positives = 67/123 (54%), Gaps = 5/123 (4%)
Query 1 TGDEWGCMCPDA-DFAWKSVAMELMLQYVKRTQGSFIENKGSALVFQYRDADPDFGQLQA 59
T EW P+ + W + + +RT S E + ++L++ Y+ AD +FG+LQA
Sbjct 673 TNGEWMTTMPEHLNMEWVDSVKHVFKYFTERTPRSHFETRDTSLIWNYKYADIEFGRLQA 732
Query 60 KDLSSHL-SELLFGYPVTVVSGKGYVEVKLLGVNKGRAVERILRTL---STLHGDIDFIF 115
+DL HL + + V VV G VEV+ +GV KG A++RIL + ++ ID++
Sbjct 733 RDLLQHLWTGPISNASVDVVQGSRSVEVRAVGVTKGAAIDRILGEIVHSKSMTTPIDYVL 792
Query 116 CIG 118
CIG
Sbjct 793 CIG 795
> ath:AT1G16980 ATTPS2; ATTPS2; alpha,alpha-trehalose-phosphate
synthase (UDP-forming)/ transferase, transferring glycosyl
groups; K00697 alpha,alpha-trehalose-phosphate synthase (UDP-forming)
[EC:2.4.1.15]
Length=821
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 41/123 (33%), Positives = 61/123 (49%), Gaps = 5/123 (4%)
Query 1 TGDEWGCMCP-DADFAWKSVAMELMLQYVKRTQGSFIENKGSALVFQYRDADPDFGQLQA 59
T EW P + + W + + RT S+ E ++LV+ Y AD +FG+ QA
Sbjct 586 TTGEWVTNMPQNVNLDWVDGVKNVFKYFTDRTPRSYFEASETSLVWNYEYADVEFGRAQA 645
Query 60 KDLSSHL-SELLFGYPVTVVSGKGYVEVKLLGVNKGRAVERILRTL---STLHGDIDFIF 115
+DL +L + + V VV G VEV +G KG A+ RIL + ++ IDF+F
Sbjct 646 RDLLQYLWAGPISNASVDVVRGNHSVEVHAIGETKGAAIGRILGEIVHRKSMTTPIDFVF 705
Query 116 CIG 118
C G
Sbjct 706 CSG 708
> ath:AT1G17000 ATTPS3; ATTPS3; alpha,alpha-trehalose-phosphate
synthase (UDP-forming); K00697 alpha,alpha-trehalose-phosphate
synthase (UDP-forming) [EC:2.4.1.15]
Length=783
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 39/123 (31%), Positives = 59/123 (47%), Gaps = 5/123 (4%)
Query 1 TGDEWGCMCP-DADFAWKSVAMELMLQYVKRTQGSFIENKGSALVFQYRDADPDFGQLQA 59
T +EW P + + W + + RT SF E ++LV+ Y AD +FG+ QA
Sbjct 564 TTEEWVTNMPQNMNLDWVDGLKNVFKYFTDRTPRSFFEASKTSLVWNYEYADVEFGRAQA 623
Query 60 KDLSSHL-SELLFGYPVTVVSGKGYVEVKLLGVNKGRAVERILRTL---STLHGDIDFIF 115
+DL +L + + VV GK VEV +GV K + IL + + ID++F
Sbjct 624 RDLLQYLWAGPISNASAEVVRGKYSVEVHAIGVTKEPEIGHILGEIVHKKAMTTPIDYVF 683
Query 116 CIG 118
C G
Sbjct 684 CSG 686
> ath:AT4G12430 trehalose-6-phosphate phosphatase, putative (EC:3.1.3.12)
Length=368
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/99 (29%), Positives = 51/99 (51%), Gaps = 6/99 (6%)
Query 24 MLQYVKRTQGSFIENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYP-VTVVSGKG 82
+++ +K +G+ +EN YR+ D + A+ + HL + YP + + G+
Sbjct 214 LVEKMKDIKGAKVENHKFCASVHYRNVDEKDWPIIAQRVHDHLKQ----YPRLRLTHGRK 269
Query 83 YVEVK-LLGVNKGRAVERILRTLSTLHGDIDFIFCIGDD 120
+EV+ ++ NKGRAVE +L +L + D IGDD
Sbjct 270 VLEVRPVIDWNKGRAVEFLLESLGLSNKDDLLPIYIGDD 308
> ath:AT4G22590 trehalose-6-phosphate phosphatase, putative (EC:3.1.3.12)
Length=377
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 29/99 (29%), Positives = 49/99 (49%), Gaps = 6/99 (6%)
Query 24 MLQYVKRTQGSFIENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYP-VTVVSGKG 82
+++ K +G+ +EN YR+ D L A+ + HL YP + + G+
Sbjct 223 LVEITKCIKGAKVENHKFCTSVHYRNVDEKDWPLVAQRVHDHLKR----YPRLRITHGRK 278
Query 83 YVEVK-LLGVNKGRAVERILRTLSTLHGDIDFIFCIGDD 120
+EV+ ++ NKG+AVE +L +L + D IGDD
Sbjct 279 VLEVRPVIEWNKGKAVEFLLESLGLSNNDEFLPIFIGDD 317
> ath:AT4G39770 trehalose-6-phosphate phosphatase, putative (EC:3.1.3.12)
Length=349
Score = 35.8 bits (81), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 25/99 (25%), Positives = 51/99 (51%), Gaps = 6/99 (6%)
Query 24 MLQYVKRTQGSFIENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYP-VTVVSGKG 82
+++ K T G+ +EN + +R D + DL++ + ++ YP + + G+
Sbjct 198 LVEKTKSTPGAQVENNKFCVSVHFRRVDEN----NWSDLANQVRSVMKDYPKLRLTQGRK 253
Query 83 YVEVK-LLGVNKGRAVERILRTLSTLHGDIDFIFCIGDD 120
+EV+ ++ +KG+A+E +L +L + F IGDD
Sbjct 254 VLEVRPIIKWDKGKALEFLLESLGYANCTDVFPLYIGDD 292
> ath:AT1G63970 ISPF; ISPF; 2-C-methyl-D-erythritol 2,4-cyclodiphosphate
synthase (EC:4.6.1.12); K01770 2-C-methyl-D-erythritol
2,4-cyclodiphosphate synthase [EC:4.6.1.12]
Length=223
Score = 32.7 bits (73), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 44/100 (44%), Gaps = 9/100 (9%)
Query 6 GCMCPDADFAWKSVAMELMLQYVKR---TQGSFIENKGSALVFQYRDADPDFGQLQAKDL 62
G + PD+D WK A + ++ R G I N + L+ Q P + +
Sbjct 124 GQIFPDSDPKWKGAASSVFIKEAVRLMDEAGYEIGNLDATLILQRPKISP-----HKETI 178
Query 63 SSHLSELLFGYPVTV-VSGKGYVEVKLLGVNKGRAVERIL 101
S+LS+LL P V + K + +V LG N+ A ++
Sbjct 179 RSNLSKLLGADPSVVNLKAKTHEKVDSLGENRSIAAHTVI 218
> ath:AT1G35910 trehalose-6-phosphate phosphatase, putative
Length=369
Score = 32.7 bits (73), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 26/99 (26%), Positives = 52/99 (52%), Gaps = 6/99 (6%)
Query 24 MLQYVKRTQGSFIENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYP-VTVVSGKG 82
+++ ++ +G+ +EN + YR D L A+ ++S LSE YP + + G+
Sbjct 217 LVEKMRDIEGANVENNKFCVSVHYRCVDQKDWGLVAEHVTSILSE----YPKLRLTQGRK 272
Query 83 YVEVK-LLGVNKGRAVERILRTLSTLHGDIDFIFCIGDD 120
+E++ + +KG+A+E +L +L + + IGDD
Sbjct 273 VLEIRPTIKWDKGKALEFLLESLGFANSNDVLPIYIGDD 311
> tgo:TGME49_093550 hypothetical protein
Length=899
Score = 30.8 bits (68), Expect = 0.98, Method: Composition-based stats.
Identities = 16/57 (28%), Positives = 28/57 (49%), Gaps = 5/57 (8%)
Query 56 QLQAKDLSSHLSELLFGYPVTVVSGKGYVEVKLLGVNKGRAVERILRTLSTLHGDID 112
Q + L H+ ++ Y V+ +GY E +LLG V+ + R + LH ++D
Sbjct 120 QQTERRLQRHIDDICLRYDSMVMEAQGYAESRLLG-----QVKDLQRVIWILHSELD 171
> xla:734182 ara-55; Hic-5
Length=459
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 29/64 (45%), Gaps = 0/64 (0%)
Query 38 NKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYPVTVVSGKGYVEVKLLGVNKGRAV 97
NKGS VF+ RD + D+ L ++ V+S G++EVK+ VN +
Sbjct 119 NKGSEEVFKPRDTEDPSSPRDTLDVPKALEDIPSPKSSEVMSTPGHMEVKIDQVNSDKVT 178
Query 98 ERIL 101
L
Sbjct 179 ASRL 182
> cel:T10C6.16 hypothetical protein
Length=201
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 83 YVEVKLLGVNKGRAVERILRTLSTLHGDIDFIFCI 117
Y LL + KG VERI R + LH D F+F +
Sbjct 142 YKNPYLLKLEKGYDVERIGRRATILHSDHKFLFAV 176
> cel:B0205.4 hypothetical protein
Length=399
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 25/97 (25%), Positives = 40/97 (41%), Gaps = 8/97 (8%)
Query 12 ADFAWKSVAMELMLQY-----VKRTQGSFIENKGSALVFQYRDADPDFG---QLQAKDLS 63
+DF + LM Q + R GS + + + + D + F Q +DLS
Sbjct 87 SDFGYSQGIGNLMFQVAGLLSIARETGSILLIPSTTTLRRAFDFETTFNDSIQFVGEDLS 146
Query 64 SHLSELLFGYPVTVVSGKGYVEVKLLGVNKGRAVERI 100
L+E L +T+ S Y + + N R +ERI
Sbjct 147 RQLAEDLNASKITLTSCCAYRNLSTILFNDSRIIERI 183
> dre:569025 Sb:cb37 protein-like
Length=1385
Score = 29.3 bits (64), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 13/37 (35%), Positives = 21/37 (56%), Gaps = 0/37 (0%)
Query 64 SHLSELLFGYPVTVVSGKGYVEVKLLGVNKGRAVERI 100
+HL E+ +P V+ G G V +LG GRA++ +
Sbjct 866 NHLEEVELAFPQNVIEGSGRATVSVLGDLLGRALKNL 902
> tgo:TGME49_119860 DNA polymerase epsilon catalytic subunit,
putative (EC:2.7.11.12 2.7.7.7); K02324 DNA polymerase epsilon
subunit 1 [EC:2.7.7.7]
Length=3124
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 35/72 (48%), Gaps = 6/72 (8%)
Query 28 VKRTQGSFIENKGSALVFQYRDADPDFGQLQAKDLSS--HLSELLFGYPV----TVVSGK 81
++RT+ S+ E AL+ DAD F + +DLS HL LL G P T S +
Sbjct 272 LRRTRASYFEQLKEALMDLLGDADVRFEKTFREDLSDPLHLRTLLPGGPAPDEGTNSSLR 331
Query 82 GYVEVKLLGVNK 93
+V++ V +
Sbjct 332 EFVQISFANVKQ 343
> ath:AT5G10100 trehalose-6-phosphate phosphatase, putative (EC:3.1.3.12)
Length=369
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 24/99 (24%), Positives = 49/99 (49%), Gaps = 6/99 (6%)
Query 24 MLQYVKRTQGSFIENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYP-VTVVSGKG 82
+L+ K T G+ +EN +R D + + +L + +L +P + + G+
Sbjct 218 LLEKTKSTPGAKVENHKFCASVHFRCVD----EKKWSELVLQVRSVLKKFPTLQLTQGRK 273
Query 83 YVEVK-LLGVNKGRAVERILRTLSTLHGDIDFIFCIGDD 120
E++ ++ +KG+A+E +L +L + + F IGDD
Sbjct 274 VFEIRPMIEWDKGKALEFLLESLGFGNTNNVFPVYIGDD 312
> eco:b1897 otsB, ECK1896, JW1886, otsP; trehalose-6-phosphate
phosphatase, biosynthetic (EC:3.1.3.12); K01087 trehalose-phosphatase
[EC:3.1.3.12]
Length=266
Score = 28.5 bits (62), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 20/76 (26%), Positives = 37/76 (48%), Gaps = 6/76 (7%)
Query 28 VKRTQGSFIENKGSALVFQYRDADPDFGQLQAKDLSSHLSELLFGYP-VTVVSGKGYVEV 86
+ + G+ +E KG A YR A L L+ ++++ +P + + GK VE+
Sbjct 114 IAQYPGAELEAKGMAFALHYRQAPQHEDALMT--LAQRITQI---WPQMALQQGKCVVEI 168
Query 87 KLLGVNKGRAVERILR 102
K G +KG A+ ++
Sbjct 169 KPRGTSKGEAIAAFMQ 184
Lambda K H
0.322 0.140 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2018002440
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40