bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6122_orf1
Length=113
Score E
Sequences producing significant alignments: (Bits) Value
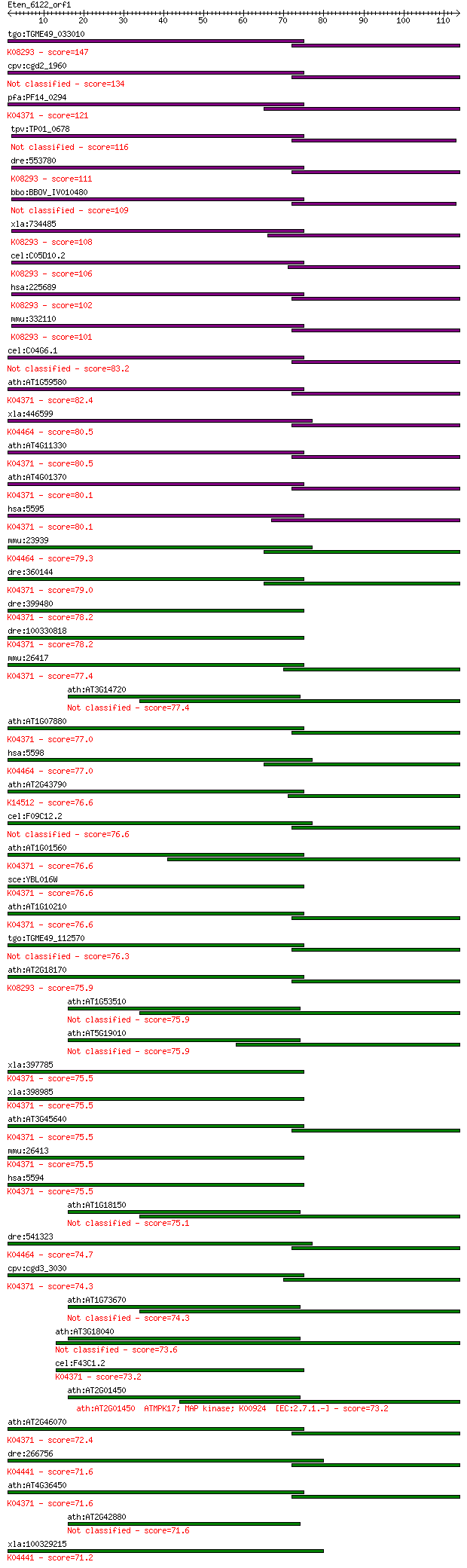
tgo:TGME49_033010 mitogen-activated protein kinase 2 (EC:2.7.1... 147 8e-36
cpv:cgd2_1960 mitogen-activated protein kinase 1, serine/threo... 134 8e-32
pfa:PF14_0294 PfMAP1; mitogen-activated protein kinase 1; K043... 121 6e-28
tpv:TP01_0678 serine/threonine protein kinase 116 2e-26
dre:553780 mapk15, MGC110345, erk7, zgc:110345; mitogen-activa... 111 5e-25
bbo:BBOV_IV010480 23.m06413; mitogen activated protein kinase ... 109 2e-24
xla:734485 mapk15, MGC99048; mitogen-activated protein kinase ... 108 4e-24
cel:C05D10.2 hypothetical protein; K08293 mitogen-activated pr... 106 2e-23
hsa:225689 MAPK15, ERK7, ERK8; mitogen-activated protein kinas... 102 4e-22
mmu:332110 Mapk15, BC048082, MGC56865, MGC56903; mitogen-activ... 101 7e-22
cel:C04G6.1 mpk-2; MAP Kinase family member (mpk-2) 83.2 2e-16
ath:AT1G59580 ATMPK2; ATMPK2 (ARABIDOPSIS THALIANA MITOGEN-ACT... 82.4 3e-16
xla:446599 mapk7, MGC81757, bmk1, erk4, prkm7; mitogen-activat... 80.5 1e-15
ath:AT4G11330 ATMPK5; ATMPK5 (MAP KINASE 5); MAP kinase/ kinas... 80.5 1e-15
ath:AT4G01370 ATMPK4; ATMPK4 (ARABIDOPSIS THALIANA MAP KINASE ... 80.1 1e-15
hsa:5595 MAPK3, ERK1, HS44KDAP, HUMKER1A, MGC20180, P44ERK1, P... 80.1 2e-15
mmu:23939 Mapk7, BMK1, ERK5, Erk5-T, PRKM7; mitogen-activated ... 79.3 2e-15
dre:360144 mapk1, zERK2; mitogen-activated protein kinase 1 (E... 79.0 3e-15
dre:399480 mapk3, ERK1, fi06b09, wu:fi06b09, zERK1; mitogen-ac... 78.2 6e-15
dre:100330818 mitogen-activated protein kinase 3-like; K04371 ... 78.2 6e-15
mmu:26417 Mapk3, Erk-1, Erk1, Ert2, Esrk1, Mnk1, Mtap2k, Prkm3... 77.4 1e-14
ath:AT3G14720 MPK19; ATMPK19; MAP kinase 77.4 1e-14
ath:AT1G07880 ATMPK13; MAP kinase/ kinase; K04371 extracellula... 77.0 1e-14
hsa:5598 MAPK7, BMK1, ERK4, ERK5, PRKM7; mitogen-activated pro... 77.0 1e-14
ath:AT2G43790 ATMPK6; ATMPK6 (ARABIDOPSIS THALIANA MAP KINASE ... 76.6 2e-14
cel:F09C12.2 hypothetical protein 76.6 2e-14
ath:AT1G01560 ATMPK11; MAP kinase/ kinase; K04371 extracellula... 76.6 2e-14
sce:YBL016W FUS3, DAC2; Fus3p (EC:2.7.11.24); K04371 extracell... 76.6 2e-14
ath:AT1G10210 ATMPK1; ATMPK1 (MITOGEN-ACTIVATED PROTEIN KINASE... 76.6 2e-14
tgo:TGME49_112570 mitogen-activated protein kinase, putative (... 76.3 2e-14
ath:AT2G18170 ATMPK7; ATMPK7 (ARABIDOPSIS THALIANA MAP KINASE ... 75.9 3e-14
ath:AT1G53510 ATMPK18; MAP kinase 75.9 3e-14
ath:AT5G19010 MPK16; MPK16; MAP kinase 75.9 3e-14
xla:397785 mapk1-b, Xp42, erk, erk2, mapk, mapk1b, mpk1; mitog... 75.5 4e-14
xla:398985 mapk1-a, MGC68934, erk, erk2, mapk, mapk1, mapk1a, ... 75.5 4e-14
ath:AT3G45640 ATMPK3; ATMPK3 (ARABIDOPSIS THALIANA MITOGEN-ACT... 75.5 4e-14
mmu:26413 Mapk1, 9030612K14Rik, AA407128, AU018647, C78273, ER... 75.5 4e-14
hsa:5594 MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2,... 75.5 4e-14
ath:AT1G18150 ATMPK8; ATMPK8; MAP kinase 75.1 5e-14
dre:541323 mapk7, bmk1, erk5, wu:fb73b02, zgc:113111; mitogen-... 74.7 6e-14
cpv:cgd3_3030 MAPK ; K04371 extracellular signal-regulated kin... 74.3 9e-14
ath:AT1G73670 ATMPK15; MAP kinase/ kinase 74.3 9e-14
ath:AT3G18040 MPK9; MPK9 (MAP KINASE 9); MAP kinase 73.6 1e-13
cel:F43C1.2 mpk-1; MAP Kinase family member (mpk-1); K04371 ex... 73.2 2e-13
ath:AT2G01450 ATMPK17; MAP kinase; K00924 [EC:2.7.1.-] 73.2 2e-13
ath:AT2G46070 MPK12; MPK12 (MITOGEN-ACTIVATED PROTEIN KINASE 1... 72.4 4e-13
dre:266756 mapk14b, MAPK14, p38b, zp38b; mitogen-activated pro... 71.6 6e-13
ath:AT4G36450 ATMPK14 (Mitogen-activated protein kinase 14); M... 71.6 6e-13
ath:AT2G42880 ATMPK20; MAP kinase 71.6 6e-13
xla:100329215 p38-beta; mitogen-activated protein kinase p38 b... 71.2 7e-13
> tgo:TGME49_033010 mitogen-activated protein kinase 2 (EC:2.7.11.24);
K08293 mitogen-activated protein kinase [EC:2.7.11.24]
Length=669
Score = 147 bits (371), Expect = 8e-36, Method: Composition-based stats.
Identities = 67/74 (90%), Positives = 71/74 (95%), Gaps = 0/74 (0%)
Query 1 ENIVRLMNVLKADNDRDIYLVFDFMETDLHAVIRAQILEAIHKQYIIYQLLRAIKYMHSG 60
ENIVRL NVLKADND+DIYLVFD+METDLHAVIRA ILE IHKQYI+YQLLRAIKYMHSG
Sbjct 71 ENIVRLKNVLKADNDKDIYLVFDYMETDLHAVIRADILEEIHKQYIVYQLLRAIKYMHSG 130
Query 61 ELLHRDMKPSNILL 74
ELLHRDMKPSN+LL
Sbjct 131 ELLHRDMKPSNVLL 144
Score = 79.7 bits (195), Expect = 2e-15, Method: Composition-based stats.
Identities = 36/42 (85%), Positives = 39/42 (92%), Gaps = 0/42 (0%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+ LKKIF+AF NATDAQRTFREIMFLQ+LAGHENIVRL NVL
Sbjct 39 VALKKIFDAFQNATDAQRTFREIMFLQELAGHENIVRLKNVL 80
> cpv:cgd2_1960 mitogen-activated protein kinase 1, serine/threonine
protein kinase
Length=710
Score = 134 bits (337), Expect = 8e-32, Method: Composition-based stats.
Identities = 59/74 (79%), Positives = 69/74 (93%), Gaps = 0/74 (0%)
Query 1 ENIVRLMNVLKADNDRDIYLVFDFMETDLHAVIRAQILEAIHKQYIIYQLLRAIKYMHSG 60
ENIV L+NVL+ADNDRD+YLVFD+METDLHAVIRA ILE +HKQY++YQL++ IKY+HSG
Sbjct 71 ENIVNLLNVLRADNDRDVYLVFDYMETDLHAVIRANILEPVHKQYVVYQLIKVIKYLHSG 130
Query 61 ELLHRDMKPSNILL 74
LLHRDMKPSNILL
Sbjct 131 GLLHRDMKPSNILL 144
Score = 68.2 bits (165), Expect = 6e-12, Method: Composition-based stats.
Identities = 30/42 (71%), Positives = 37/42 (88%), Gaps = 0/42 (0%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+ +KKIF+AF N+TDAQRTFREIM L +L+GHENIV L+NVL
Sbjct 39 VAVKKIFDAFQNSTDAQRTFREIMILTELSGHENIVNLLNVL 80
> pfa:PF14_0294 PfMAP1; mitogen-activated protein kinase 1; K04371
extracellular signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=914
Score = 121 bits (303), Expect = 6e-28, Method: Composition-based stats.
Identities = 55/74 (74%), Positives = 68/74 (91%), Gaps = 0/74 (0%)
Query 1 ENIVRLMNVLKADNDRDIYLVFDFMETDLHAVIRAQILEAIHKQYIIYQLLRAIKYMHSG 60
+NI++LM+V+KA ND DIYL+FDFMETDLH VI+A +LE IHK+YIIYQLLRA+KY+HSG
Sbjct 81 DNIIKLMDVIKAKNDNDIYLIFDFMETDLHEVIKADLLEEIHKKYIIYQLLRALKYIHSG 140
Query 61 ELLHRDMKPSNILL 74
LLHRD+KPSNIL+
Sbjct 141 GLLHRDIKPSNILV 154
Score = 62.0 bits (149), Expect = 4e-10, Method: Composition-based stats.
Identities = 31/50 (62%), Positives = 40/50 (80%), Gaps = 1/50 (2%)
Query 65 RDMKPSNIL-LKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
R K NI+ +KKIF AF N TDAQRTFREI+FL +L GH+NI++LM+V+
Sbjct 41 RCKKNKNIVAVKKIFGAFQNCTDAQRTFREIIFLYELNGHDNIIKLMDVI 90
> tpv:TP01_0678 serine/threonine protein kinase
Length=677
Score = 116 bits (290), Expect = 2e-26, Method: Composition-based stats.
Identities = 50/73 (68%), Positives = 67/73 (91%), Gaps = 0/73 (0%)
Query 2 NIVRLMNVLKADNDRDIYLVFDFMETDLHAVIRAQILEAIHKQYIIYQLLRAIKYMHSGE 61
NIV+L +V ADN+RD+YLVF+++ETDLHAVIR+ ILE +HK+YI+YQLL+AI ++H+G+
Sbjct 74 NIVKLRDVYPADNNRDVYLVFEYVETDLHAVIRSNILEEVHKRYILYQLLKAIHFIHTGD 133
Query 62 LLHRDMKPSNILL 74
LLHRD+KPSNILL
Sbjct 134 LLHRDLKPSNILL 146
Score = 58.5 bits (140), Expect = 5e-09, Method: Composition-based stats.
Identities = 27/41 (65%), Positives = 34/41 (82%), Gaps = 0/41 (0%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNV 112
+ LKKIF+AF N+TDAQRT+REIMFLQ+L NIV+L +V
Sbjct 41 VALKKIFDAFRNSTDAQRTYREIMFLQKLKKCPNIVKLRDV 81
> dre:553780 mapk15, MGC110345, erk7, zgc:110345; mitogen-activated
protein kinase 15 (EC:2.7.11.24); K08293 mitogen-activated
protein kinase [EC:2.7.11.24]
Length=524
Score = 111 bits (278), Expect = 5e-25, Method: Composition-based stats.
Identities = 47/74 (63%), Positives = 66/74 (89%), Gaps = 1/74 (1%)
Query 2 NIVRLMNVLKADNDRDIYLVFDFMETDLHAVIR-AQILEAIHKQYIIYQLLRAIKYMHSG 60
NI++L+NV++A ND+DIYL+F+FM+TDLHAVI+ +L+ IHK+Y++YQLL+A KY+HSG
Sbjct 73 NIIKLLNVIRAQNDKDIYLIFEFMDTDLHAVIKKGNLLKDIHKRYVMYQLLKATKYLHSG 132
Query 61 ELLHRDMKPSNILL 74
++HRD KPSNILL
Sbjct 133 NVIHRDQKPSNILL 146
Score = 64.7 bits (156), Expect = 6e-11, Method: Composition-based stats.
Identities = 27/42 (64%), Positives = 35/42 (83%), Gaps = 0/42 (0%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+ +KKIF+AF N TDAQRTFREIMFLQ+ H NI++L+NV+
Sbjct 40 VAVKKIFDAFRNRTDAQRTFREIMFLQEFGDHPNIIKLLNVI 81
> bbo:BBOV_IV010480 23.m06413; mitogen activated protein kinase
(EC:2.7.1.-)
Length=506
Score = 109 bits (273), Expect = 2e-24, Method: Composition-based stats.
Identities = 48/73 (65%), Positives = 63/73 (86%), Gaps = 0/73 (0%)
Query 2 NIVRLMNVLKADNDRDIYLVFDFMETDLHAVIRAQILEAIHKQYIIYQLLRAIKYMHSGE 61
NI+ L +V A NDRD+YL F++++TDLH+VIR ILE IHK+YIIYQLL+AI ++HSG+
Sbjct 72 NIIELKDVYPAKNDRDLYLTFEYIDTDLHSVIRINILEEIHKKYIIYQLLKAINFIHSGD 131
Query 62 LLHRDMKPSNILL 74
LLHRD+KPSN+LL
Sbjct 132 LLHRDLKPSNVLL 144
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 25/41 (60%), Positives = 31/41 (75%), Gaps = 0/41 (0%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNV 112
+ LKKIF+AF N+TDAQRT+REIMFL L NI+ L +V
Sbjct 39 VALKKIFDAFRNSTDAQRTYREIMFLHYLRKAHNIIELKDV 79
> xla:734485 mapk15, MGC99048; mitogen-activated protein kinase
15 (EC:2.7.11.24); K08293 mitogen-activated protein kinase
[EC:2.7.11.24]
Length=586
Score = 108 bits (270), Expect = 4e-24, Method: Composition-based stats.
Identities = 47/74 (63%), Positives = 64/74 (86%), Gaps = 1/74 (1%)
Query 2 NIVRLMNVLKADNDRDIYLVFDFMETDLHAVIR-AQILEAIHKQYIIYQLLRAIKYMHSG 60
NI++L+NV++A ND+DIYLVF+ METDLHAVI+ +L+ IH +YI+YQLL+A K++HSG
Sbjct 73 NIIKLLNVIRAQNDKDIYLVFEHMETDLHAVIKKGNLLKDIHMRYILYQLLKATKFIHSG 132
Query 61 ELLHRDMKPSNILL 74
++HRD KPSNILL
Sbjct 133 NVIHRDQKPSNILL 146
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 30/49 (61%), Positives = 38/49 (77%), Gaps = 1/49 (2%)
Query 66 DMKPSNIL-LKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
D K I+ +KKIF+AF N TDAQRTFREIMFLQ+ H NI++L+NV+
Sbjct 33 DRKSGEIVAVKKIFDAFRNRTDAQRTFREIMFLQEFGEHPNIIKLLNVI 81
> cel:C05D10.2 hypothetical protein; K08293 mitogen-activated
protein kinase [EC:2.7.11.24]
Length=470
Score = 106 bits (264), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 46/74 (62%), Positives = 62/74 (83%), Gaps = 1/74 (1%)
Query 2 NIVRLMNVLKADNDRDIYLVFDFMETDLHAVIR-AQILEAIHKQYIIYQLLRAIKYMHSG 60
N+++L N+ +ADNDRDIYL F+FME DLH VI+ IL+ +HKQYI+ QL RAI+++HSG
Sbjct 72 NVIKLYNIFRADNDRDIYLAFEFMEADLHNVIKKGSILKDVHKQYIMCQLFRAIRFLHSG 131
Query 61 ELLHRDMKPSNILL 74
+LHRD+KPSN+LL
Sbjct 132 NVLHRDLKPSNVLL 145
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 24/43 (55%), Positives = 33/43 (76%), Gaps = 0/43 (0%)
Query 71 NILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+ LKKIF+AF N TD+QRTFRE+MFLQ+ H N+++L N+
Sbjct 38 TVALKKIFDAFRNPTDSQRTFREVMFLQEFGKHPNVIKLYNIF 80
> hsa:225689 MAPK15, ERK7, ERK8; mitogen-activated protein kinase
15 (EC:2.7.11.24); K08293 mitogen-activated protein kinase
[EC:2.7.11.24]
Length=544
Score = 102 bits (253), Expect = 4e-22, Method: Composition-based stats.
Identities = 44/74 (59%), Positives = 63/74 (85%), Gaps = 1/74 (1%)
Query 2 NIVRLMNVLKADNDRDIYLVFDFMETDLHAVIR-AQILEAIHKQYIIYQLLRAIKYMHSG 60
NI+ L++V++A+NDRDIYLVF+FM+TDL+AVIR +L+ +H + I YQLLRA +++HSG
Sbjct 72 NIISLLDVIRAENDRDIYLVFEFMDTDLNAVIRKGGLLQDVHVRSIFYQLLRATRFLHSG 131
Query 61 ELLHRDMKPSNILL 74
++HRD KPSN+LL
Sbjct 132 HVVHRDQKPSNVLL 145
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 23/42 (54%), Positives = 32/42 (76%), Gaps = 0/42 (0%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+ +KKIF+AF + TDAQRTFREI LQ+ H NI+ L++V+
Sbjct 39 VAIKKIFDAFRDKTDAQRTFREITLLQEFGDHPNIISLLDVI 80
> mmu:332110 Mapk15, BC048082, MGC56865, MGC56903; mitogen-activated
protein kinase 15 (EC:2.7.11.24); K08293 mitogen-activated
protein kinase [EC:2.7.11.24]
Length=549
Score = 101 bits (251), Expect = 7e-22, Method: Composition-based stats.
Identities = 45/74 (60%), Positives = 63/74 (85%), Gaps = 1/74 (1%)
Query 2 NIVRLMNVLKADNDRDIYLVFDFMETDLHAVI-RAQILEAIHKQYIIYQLLRAIKYMHSG 60
NI+RL++V+ A NDRDIYLVF+ M+TDL+AVI + ++L+ IHK+ I YQLLRA K++HSG
Sbjct 73 NIIRLLDVIPAKNDRDIYLVFESMDTDLNAVIQKGRLLKDIHKRCIFYQLLRATKFIHSG 132
Query 61 ELLHRDMKPSNILL 74
++HRD KP+N+LL
Sbjct 133 RVIHRDQKPANVLL 146
Score = 60.8 bits (146), Expect = 1e-09, Method: Composition-based stats.
Identities = 24/42 (57%), Positives = 34/42 (80%), Gaps = 0/42 (0%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+ +KKIF+AF + DAQRTFREIM L++ GH NI+RL++V+
Sbjct 40 VAIKKIFDAFRDQIDAQRTFREIMLLKEFGGHPNIIRLLDVI 81
> cel:C04G6.1 mpk-2; MAP Kinase family member (mpk-2)
Length=605
Score = 83.2 bits (204), Expect = 2e-16, Method: Composition-based stats.
Identities = 39/77 (50%), Positives = 56/77 (72%), Gaps = 3/77 (3%)
Query 1 ENIVRLMNVLKAD--NDRDIYLVFDFMETDLHAVIRA-QILEAIHKQYIIYQLLRAIKYM 57
ENI+ ++++ A+ + +DIYLV D METDLH ++ + Q L H QY YQLLR +KY+
Sbjct 114 ENIIAVLDMFTAEGAHGKDIYLVMDLMETDLHQILHSRQTLMEQHFQYFFYQLLRGLKYL 173
Query 58 HSGELLHRDMKPSNILL 74
HS ++HRD+KPSN+LL
Sbjct 174 HSAGIIHRDLKPSNLLL 190
Score = 32.3 bits (72), Expect = 0.35, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 28/42 (66%), Gaps = 1/42 (2%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+ +KKI AF T A+R+ RE+ L++L HENI+ ++++
Sbjct 83 VAIKKIPRAFTAHTLAKRSLREVRILRELL-HENIIAVLDMF 123
> ath:AT1G59580 ATMPK2; ATMPK2 (ARABIDOPSIS THALIANA MITOGEN-ACTIVATED
PROTEIN KINASE HOMOLOG 2); MAP kinase/ kinase/ protein
kinase; K04371 extracellular signal-regulated kinase 1/2
[EC:2.7.11.24]
Length=376
Score = 82.4 bits (202), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 38/78 (48%), Positives = 59/78 (75%), Gaps = 4/78 (5%)
Query 1 ENIVRLMNVLKADNDR---DIYLVFDFMETDLHAVIRA-QILEAIHKQYIIYQLLRAIKY 56
EN+V L +V+ A++ R D+YLV++ M+TDLH +I++ Q+L H QY ++QLLR +KY
Sbjct 89 ENVVALKDVMMANHKRSFKDVYLVYELMDTDLHQIIKSSQVLSNDHCQYFLFQLLRGLKY 148
Query 57 MHSGELLHRDMKPSNILL 74
+HS +LHRD+KP N+L+
Sbjct 149 IHSANILHRDLKPGNLLV 166
Score = 35.4 bits (80), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 19/42 (45%), Positives = 26/42 (61%), Gaps = 1/42 (2%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+ +KKI F N DA RT RE+ L+ L HEN+V L +V+
Sbjct 58 VAIKKIHNVFENRIDALRTLRELKLLRHLR-HENVVALKDVM 98
> xla:446599 mapk7, MGC81757, bmk1, erk4, prkm7; mitogen-activated
protein kinase 7; K04464 mitogen-activated protein kinase
7 [EC:2.7.11.24]
Length=925
Score = 80.5 bits (197), Expect = 1e-15, Method: Composition-based stats.
Identities = 37/81 (45%), Positives = 57/81 (70%), Gaps = 5/81 (6%)
Query 1 ENIVRLMNVLKAD---ND-RDIYLVFDFMETDLHAVIRA-QILEAIHKQYIIYQLLRAIK 55
+N++ + ++LK ND R +Y+V D ME+DLH +I + Q L H +Y +YQLLR +K
Sbjct 107 DNVIAIKDILKPSVPYNDFRSVYVVLDLMESDLHQIIHSSQPLTLEHARYFLYQLLRGLK 166
Query 56 YMHSGELLHRDMKPSNILLKK 76
Y+HS +LHRD+KPSN+L+ +
Sbjct 167 YIHSANVLHRDLKPSNLLINE 187
Score = 33.9 bits (76), Expect = 0.13, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 27/42 (64%), Gaps = 1/42 (2%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+ +KKI AF T+A+RT RE+ L+ H+N++ + ++L
Sbjct 76 VAIKKIPNAFDVVTNAKRTLRELKILKHFK-HDNVIAIKDIL 116
> ath:AT4G11330 ATMPK5; ATMPK5 (MAP KINASE 5); MAP kinase/ kinase;
K04371 extracellular signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=376
Score = 80.5 bits (197), Expect = 1e-15, Method: Composition-based stats.
Identities = 38/78 (48%), Positives = 58/78 (74%), Gaps = 4/78 (5%)
Query 1 ENIVRLMNVL---KADNDRDIYLVFDFMETDLHAVIRA-QILEAIHKQYIIYQLLRAIKY 56
EN+V + +++ K ++ D+Y+VF+ M+TDLH +IR+ Q L H QY +YQ+LR +KY
Sbjct 100 ENVVVIKDIIRPPKKEDFVDVYIVFELMDTDLHQIIRSNQSLNDDHCQYFLYQILRGLKY 159
Query 57 MHSGELLHRDMKPSNILL 74
+HS +LHRD+KPSN+LL
Sbjct 160 IHSANVLHRDLKPSNLLL 177
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 20/42 (47%), Positives = 29/42 (69%), Gaps = 1/42 (2%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
I +KKI +AF N DA+RT REI L+ L HEN+V + +++
Sbjct 69 IAIKKIGKAFDNKVDAKRTLREIKLLRHLE-HENVVVIKDII 109
> ath:AT4G01370 ATMPK4; ATMPK4 (ARABIDOPSIS THALIANA MAP KINASE
4); MAP kinase/ kinase; K04371 extracellular signal-regulated
kinase 1/2 [EC:2.7.11.24]
Length=376
Score = 80.1 bits (196), Expect = 1e-15, Method: Composition-based stats.
Identities = 36/78 (46%), Positives = 58/78 (74%), Gaps = 4/78 (5%)
Query 1 ENIVRLMNVLKA---DNDRDIYLVFDFMETDLHAVIRA-QILEAIHKQYIIYQLLRAIKY 56
EN++ + +++K +N D+Y+V++ M+TDLH +IR+ Q L H ++ +YQLLR +KY
Sbjct 100 ENVIAVKDIIKPPQRENFNDVYIVYELMDTDLHQIIRSNQPLTDDHCRFFLYQLLRGLKY 159
Query 57 MHSGELLHRDMKPSNILL 74
+HS +LHRD+KPSN+LL
Sbjct 160 VHSANVLHRDLKPSNLLL 177
Score = 38.1 bits (87), Expect = 0.008, Method: Composition-based stats.
Identities = 17/42 (40%), Positives = 28/42 (66%), Gaps = 1/42 (2%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+ +KKI AF N DA+RT REI L+ + HEN++ + +++
Sbjct 69 VAIKKIGNAFDNIIDAKRTLREIKLLKHM-DHENVIAVKDII 109
> hsa:5595 MAPK3, ERK1, HS44KDAP, HUMKER1A, MGC20180, P44ERK1,
P44MAPK, PRKM3; mitogen-activated protein kinase 3 (EC:2.7.11.24);
K04371 extracellular signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=357
Score = 80.1 bits (196), Expect = 2e-15, Method: Composition-based stats.
Identities = 35/77 (45%), Positives = 57/77 (74%), Gaps = 3/77 (3%)
Query 1 ENIVRLMNVLKA---DNDRDIYLVFDFMETDLHAVIRAQILEAIHKQYIIYQLLRAIKYM 57
EN++ + ++L+A + RD+Y+V D METDL+ ++++Q L H Y +YQ+LR +KY+
Sbjct 98 ENVIGIRDILRASTLEAMRDVYIVQDLMETDLYKLLKSQQLSNDHICYFLYQILRGLKYI 157
Query 58 HSGELLHRDMKPSNILL 74
HS +LHRD+KPSN+L+
Sbjct 158 HSANVLHRDLKPSNLLI 174
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 28/47 (59%), Gaps = 2/47 (4%)
Query 67 MKPSNILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
++ + + +KKI F + T QRT REI L + HEN++ + ++L
Sbjct 63 VRKTRVAIKKI-SPFEHQTYCQRTLREIQILLRFR-HENVIGIRDIL 107
> mmu:23939 Mapk7, BMK1, ERK5, Erk5-T, PRKM7; mitogen-activated
protein kinase 7 (EC:2.7.11.24); K04464 mitogen-activated
protein kinase 7 [EC:2.7.11.24]
Length=806
Score = 79.3 bits (194), Expect = 2e-15, Method: Composition-based stats.
Identities = 36/81 (44%), Positives = 56/81 (69%), Gaps = 5/81 (6%)
Query 1 ENIVRLMNVLKAD----NDRDIYLVFDFMETDLHAVIRA-QILEAIHKQYIIYQLLRAIK 55
+NI+ + ++LK R +Y+V D ME+DLH +I + Q L H +Y +YQLLR +K
Sbjct 112 DNIIAIKDILKPTVPYGEFRSVYVVLDLMESDLHQIIHSSQPLTLEHVRYFLYQLLRGLK 171
Query 56 YMHSGELLHRDMKPSNILLKK 76
YMHS +++HRD+KPSN+L+ +
Sbjct 172 YMHSAQVIHRDLKPSNLLVNE 192
Score = 35.8 bits (81), Expect = 0.035, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 29/49 (59%), Gaps = 1/49 (2%)
Query 65 RDMKPSNILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
R + + +KKI AF T+A+RT RE+ L+ H+NI+ + ++L
Sbjct 74 RRLTGQQVAIKKIPNAFDVVTNAKRTLRELKILKHFK-HDNIIAIKDIL 121
> dre:360144 mapk1, zERK2; mitogen-activated protein kinase 1
(EC:2.7.11.24); K04371 extracellular signal-regulated kinase
1/2 [EC:2.7.11.24]
Length=369
Score = 79.0 bits (193), Expect = 3e-15, Method: Composition-based stats.
Identities = 35/77 (45%), Positives = 55/77 (71%), Gaps = 3/77 (3%)
Query 1 ENIVRLMNVLKA---DNDRDIYLVFDFMETDLHAVIRAQILEAIHKQYIIYQLLRAIKYM 57
ENI+ + ++++ D +D+Y+V D METDL+ +++ Q L H Y +YQ+LR +KY+
Sbjct 90 ENIIGINDIIRTPTIDQMKDVYIVQDLMETDLYKLLKTQHLSNDHICYFLYQILRGLKYI 149
Query 58 HSGELLHRDMKPSNILL 74
HS +LHRD+KPSN+LL
Sbjct 150 HSANVLHRDLKPSNLLL 166
Score = 28.1 bits (61), Expect = 7.2, Method: Composition-based stats.
Identities = 19/49 (38%), Positives = 28/49 (57%), Gaps = 3/49 (6%)
Query 65 RDMKPSNILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
RD K + +KKI F + T QRT REI L + HENI+ + +++
Sbjct 54 RDNK-VRVAIKKI-SPFEHQTYCQRTLREIKILLRFK-HENIIGINDII 99
> dre:399480 mapk3, ERK1, fi06b09, wu:fi06b09, zERK1; mitogen-activated
protein kinase 3 (EC:2.7.11.1); K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=392
Score = 78.2 bits (191), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 37/77 (48%), Positives = 56/77 (72%), Gaps = 3/77 (3%)
Query 1 ENIVRLMNVLKA---DNDRDIYLVFDFMETDLHAVIRAQILEAIHKQYIIYQLLRAIKYM 57
ENI+ + ++L+A D RD+Y+V D METDL+ +++ Q L H Y +YQ+LR +KY+
Sbjct 112 ENIIGINDILRARHIDYMRDVYIVQDLMETDLYKLLKTQQLSNDHICYFLYQILRGLKYI 171
Query 58 HSGELLHRDMKPSNILL 74
HS +LHRD+KPSN+L+
Sbjct 172 HSANVLHRDLKPSNLLI 188
> dre:100330818 mitogen-activated protein kinase 3-like; K04371
extracellular signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=392
Score = 78.2 bits (191), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 37/77 (48%), Positives = 56/77 (72%), Gaps = 3/77 (3%)
Query 1 ENIVRLMNVLKA---DNDRDIYLVFDFMETDLHAVIRAQILEAIHKQYIIYQLLRAIKYM 57
ENI+ + ++L+A D RD+Y+V D METDL+ +++ Q L H Y +YQ+LR +KY+
Sbjct 112 ENIIGINDILRARHIDYMRDVYIVQDLMETDLYKLLKTQQLSNDHICYFLYQILRGLKYI 171
Query 58 HSGELLHRDMKPSNILL 74
HS +LHRD+KPSN+L+
Sbjct 172 HSANVLHRDLKPSNLLI 188
> mmu:26417 Mapk3, Erk-1, Erk1, Ert2, Esrk1, Mnk1, Mtap2k, Prkm3,
p44, p44erk1, p44mapk; mitogen-activated protein kinase
3 (EC:2.7.11.24); K04371 extracellular signal-regulated kinase
1/2 [EC:2.7.11.24]
Length=380
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 35/77 (45%), Positives = 56/77 (72%), Gaps = 3/77 (3%)
Query 1 ENIVRLMNVLKADN---DRDIYLVFDFMETDLHAVIRAQILEAIHKQYIIYQLLRAIKYM 57
EN++ + ++L+A RD+Y+V D METDL+ ++++Q L H Y +YQ+LR +KY+
Sbjct 99 ENVIGIRDILRAPTLEAMRDVYIVQDLMETDLYKLLKSQQLSNDHICYFLYQILRGLKYI 158
Query 58 HSGELLHRDMKPSNILL 74
HS +LHRD+KPSN+L+
Sbjct 159 HSANVLHRDLKPSNLLI 175
Score = 28.5 bits (62), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 26/44 (59%), Gaps = 2/44 (4%)
Query 70 SNILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+ + +KKI F + T QRT REI L + HEN++ + ++L
Sbjct 67 TRVAIKKI-SPFEHQTYCQRTLREIQILLRFR-HENVIGIRDIL 108
> ath:AT3G14720 MPK19; ATMPK19; MAP kinase
Length=598
Score = 77.4 bits (189), Expect = 1e-14, Method: Composition-based stats.
Identities = 33/59 (55%), Positives = 45/59 (76%), Gaps = 1/59 (1%)
Query 16 RDIYLVFDFMETDLHAVIRAQI-LEAIHKQYIIYQLLRAIKYMHSGELLHRDMKPSNIL 73
+DIY+VF+ ME+DLH VI+A L H Q+ +YQ+LRA+KYMH+ + HRD+KP NIL
Sbjct 100 KDIYVVFELMESDLHQVIKANDDLTREHHQFFLYQMLRALKYMHTANVYHRDLKPKNIL 158
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 24/80 (30%), Positives = 40/80 (50%), Gaps = 12/80 (15%)
Query 34 RAQILEAIHKQYIIYQLLRAIKYMHSGELLHRDMKPSNILLKKIFEAFHNATDAQRTFRE 93
R +ILE I K Y ++ A +GE + +KKI + F + +DA R RE
Sbjct 24 RYRILEVIGKGS--YGVVCAAIDTQTGE---------KVAIKKINDVFEHVSDALRILRE 72
Query 94 IMFLQQLAGHENIVRLMNVL 113
+ L+ L H +IV + +++
Sbjct 73 VKLLRLLR-HPDIVEIKSIM 91
> ath:AT1G07880 ATMPK13; MAP kinase/ kinase; K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=363
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 34/78 (43%), Positives = 60/78 (76%), Gaps = 4/78 (5%)
Query 1 ENIVRLMNVLK-ADNDR--DIYLVFDFMETDLHAVIRA-QILEAIHKQYIIYQLLRAIKY 56
+N++++ ++++ + +R D+Y+V++ M+TDLH +IR+ Q L H QY +YQ+LR +KY
Sbjct 90 DNVIKIKDIIELPEKERFEDVYIVYELMDTDLHQIIRSTQTLTDDHCQYFLYQILRGLKY 149
Query 57 MHSGELLHRDMKPSNILL 74
+HS +LHRD+KPSN++L
Sbjct 150 IHSANVLHRDLKPSNLVL 167
Score = 37.0 bits (84), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 28/42 (66%), Gaps = 1/42 (2%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+ +KKI AF N DA+RT REI L + H+N++++ +++
Sbjct 59 VAIKKIANAFDNRVDAKRTLREIKLLSHM-DHDNVIKIKDII 99
> hsa:5598 MAPK7, BMK1, ERK4, ERK5, PRKM7; mitogen-activated protein
kinase 7 (EC:2.7.11.24); K04464 mitogen-activated protein
kinase 7 [EC:2.7.11.24]
Length=816
Score = 77.0 bits (188), Expect = 1e-14, Method: Composition-based stats.
Identities = 34/81 (41%), Positives = 56/81 (69%), Gaps = 5/81 (6%)
Query 1 ENIVRLMNVLKAD----NDRDIYLVFDFMETDLHAVIRA-QILEAIHKQYIIYQLLRAIK 55
+NI+ + ++L+ + +Y+V D ME+DLH +I + Q L H +Y +YQLLR +K
Sbjct 112 DNIIAIKDILRPTVPYGEFKSVYVVLDLMESDLHQIIHSSQPLTLEHVRYFLYQLLRGLK 171
Query 56 YMHSGELLHRDMKPSNILLKK 76
YMHS +++HRD+KPSN+L+ +
Sbjct 172 YMHSAQVIHRDLKPSNLLVNE 192
Score = 35.8 bits (81), Expect = 0.035, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 29/49 (59%), Gaps = 1/49 (2%)
Query 65 RDMKPSNILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
R + + +KKI AF T+A+RT RE+ L+ H+NI+ + ++L
Sbjct 74 RRLTGQQVAIKKIPNAFDVVTNAKRTLRELKILKHFK-HDNIIAIKDIL 121
> ath:AT2G43790 ATMPK6; ATMPK6 (ARABIDOPSIS THALIANA MAP KINASE
6); MAP kinase/ kinase; K14512 mitogen-activated protein kinase
6 [EC:2.7.11.24]
Length=395
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 36/78 (46%), Positives = 55/78 (70%), Gaps = 4/78 (5%)
Query 1 ENIVRLMNVLKA---DNDRDIYLVFDFMETDLHAVIRA-QILEAIHKQYIIYQLLRAIKY 56
ENIV + +++ + D+Y+ ++ M+TDLH +IR+ Q L H QY +YQ+LR +KY
Sbjct 120 ENIVAIRDIIPPPLRNAFNDVYIAYELMDTDLHQIIRSNQALSEEHCQYFLYQILRGLKY 179
Query 57 MHSGELLHRDMKPSNILL 74
+HS +LHRD+KPSN+LL
Sbjct 180 IHSANVLHRDLKPSNLLL 197
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 19/43 (44%), Positives = 29/43 (67%), Gaps = 1/43 (2%)
Query 71 NILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
++ +KKI AF N DA+RT REI L+ + HENIV + +++
Sbjct 88 SVAIKKIANAFDNKIDAKRTLREIKLLRHM-DHENIVAIRDII 129
> cel:F09C12.2 hypothetical protein
Length=418
Score = 76.6 bits (187), Expect = 2e-14, Method: Composition-based stats.
Identities = 36/79 (45%), Positives = 57/79 (72%), Gaps = 3/79 (3%)
Query 1 ENIVRLMNVL--KADNDRDIYLVFDFMETDLHAVIRA-QILEAIHKQYIIYQLLRAIKYM 57
ENI+ +VL ++ + +DIY+V D METDL +V+++ Q L H QY YQ+L+ +KY+
Sbjct 101 ENIINSTDVLMRESGSGQDIYIVMDLMETDLLSVLKSNQTLNEKHFQYFFYQILKGLKYL 160
Query 58 HSGELLHRDMKPSNILLKK 76
HS ++HRD+KP+N+LL +
Sbjct 161 HSAGIIHRDLKPANLLLNE 179
Score = 30.8 bits (68), Expect = 1.0, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+ +KKI F N + A+ REI ++L+ HENI+ +VL
Sbjct 70 VAIKKITRVFKNQSTAKCALREIRITRELS-HENIINSTDVL 110
> ath:AT1G01560 ATMPK11; MAP kinase/ kinase; K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=369
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 34/78 (43%), Positives = 59/78 (75%), Gaps = 4/78 (5%)
Query 1 ENIVRLMNVLKA---DNDRDIYLVFDFMETDLHAVIRA-QILEAIHKQYIIYQLLRAIKY 56
+N++ ++++++ DN D+++V++ M+TDLH +IR+ Q L H ++ +YQLLR +KY
Sbjct 97 DNVIAIIDIIRPPQPDNFNDVHIVYELMDTDLHHIIRSNQPLTDDHSRFFLYQLLRGLKY 156
Query 57 MHSGELLHRDMKPSNILL 74
+HS +LHRD+KPSN+LL
Sbjct 157 VHSANVLHRDLKPSNLLL 174
Score = 36.2 bits (82), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 40/75 (53%), Gaps = 5/75 (6%)
Query 41 IHKQYIIYQLLRAIKYMHSGELL--HRDMKPSNILLKKIFEAFHNATDAQRTFREIMFLQ 98
+ K+Y+ LR I SG + + +KKI AF N DA+RT REI L+
Sbjct 35 VSKKYV--PPLRPIGRGASGIVCAAWNSETGEEVAIKKIGNAFGNIIDAKRTLREIKLLK 92
Query 99 QLAGHENIVRLMNVL 113
+ H+N++ +++++
Sbjct 93 HM-DHDNVIAIIDII 106
> sce:YBL016W FUS3, DAC2; Fus3p (EC:2.7.11.24); K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=353
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 35/77 (45%), Positives = 53/77 (68%), Gaps = 3/77 (3%)
Query 1 ENIVRLMNVLKAD---NDRDIYLVFDFMETDLHAVIRAQILEAIHKQYIIYQLLRAIKYM 57
ENI+ + N+ + D N ++Y++ + M+TDLH VI Q+L H QY IYQ LRA+K +
Sbjct 69 ENIITIFNIQRPDSFENFNEVYIIQELMQTDLHRVISTQMLSDDHIQYFIYQTLRAVKVL 128
Query 58 HSGELLHRDMKPSNILL 74
H ++HRD+KPSN+L+
Sbjct 129 HGSNVIHRDLKPSNLLI 145
> ath:AT1G10210 ATMPK1; ATMPK1 (MITOGEN-ACTIVATED PROTEIN KINASE
1); MAP kinase/ kinase; K04371 extracellular signal-regulated
kinase 1/2 [EC:2.7.11.24]
Length=370
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 35/78 (44%), Positives = 57/78 (73%), Gaps = 4/78 (5%)
Query 1 ENIVRLMNVLKADND---RDIYLVFDFMETDLHAVIRA-QILEAIHKQYIIYQLLRAIKY 56
EN++ L +V+ + +D+YLV++ M+TDLH +I++ Q+L H QY ++QLLR +KY
Sbjct 89 ENVIALKDVMMPIHKMSFKDVYLVYELMDTDLHQIIKSSQVLSNDHCQYFLFQLLRGLKY 148
Query 57 MHSGELLHRDMKPSNILL 74
+HS +LHRD+KP N+L+
Sbjct 149 IHSANILHRDLKPGNLLV 166
Score = 33.5 bits (75), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 26/42 (61%), Gaps = 1/42 (2%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+ +KKI + N DA RT RE+ L+ L HEN++ L +V+
Sbjct 58 VAIKKIHNVYENRIDALRTLRELKLLRHLR-HENVIALKDVM 98
> tgo:TGME49_112570 mitogen-activated protein kinase, putative
(EC:2.7.11.24)
Length=1212
Score = 76.3 bits (186), Expect = 2e-14, Method: Composition-based stats.
Identities = 37/77 (48%), Positives = 52/77 (67%), Gaps = 3/77 (3%)
Query 1 ENIVRLMNVLK--ADNDRDIYLVFDFMETDLHAVIRA-QILEAIHKQYIIYQLLRAIKYM 57
ENI+ L+ +L + DIYLV D M+TDLH VI + Q L H QY +YQLL + ++
Sbjct 168 ENIINLVEILDPLTPDFEDIYLVSDLMDTDLHRVIYSRQPLTPEHHQYFLYQLLLGLSFL 227
Query 58 HSGELLHRDMKPSNILL 74
H +++HRD+KPSNIL+
Sbjct 228 HQADIIHRDLKPSNILV 244
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 18/42 (42%), Positives = 29/42 (69%), Gaps = 1/42 (2%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+ +KKI + F + DA+R +REI L++L HENI+ L+ +L
Sbjct 137 VAVKKIGDLFRDLIDAKRIYREIKILKELK-HENIINLVEIL 177
> ath:AT2G18170 ATMPK7; ATMPK7 (ARABIDOPSIS THALIANA MAP KINASE
7); MAP kinase/ kinase; K08293 mitogen-activated protein kinase
[EC:2.7.11.24]
Length=368
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 35/78 (44%), Positives = 56/78 (71%), Gaps = 4/78 (5%)
Query 1 ENIVRLMNVLKADND---RDIYLVFDFMETDLHAVIRA-QILEAIHKQYIIYQLLRAIKY 56
EN++ L +V+ N +D+YLV++ M+TDLH +I++ Q L H +Y ++QLLR +KY
Sbjct 89 ENVIALKDVMLPANRSSFKDVYLVYELMDTDLHQIIKSSQSLSDDHCKYFLFQLLRGLKY 148
Query 57 MHSGELLHRDMKPSNILL 74
+HS +LHRD+KP N+L+
Sbjct 149 LHSANILHRDLKPGNLLV 166
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 26/42 (61%), Gaps = 1/42 (2%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+ +KKI F N DA RT RE+ L+ + HEN++ L +V+
Sbjct 58 VAIKKIHNVFENRVDALRTLRELKLLRHVR-HENVIALKDVM 98
> ath:AT1G53510 ATMPK18; MAP kinase
Length=615
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 32/59 (54%), Positives = 45/59 (76%), Gaps = 1/59 (1%)
Query 16 RDIYLVFDFMETDLHAVIRAQI-LEAIHKQYIIYQLLRAIKYMHSGELLHRDMKPSNIL 73
+DIY+VF+ ME+DLH VI+A L H Q+ +YQ+LRA+K+MH+ + HRD+KP NIL
Sbjct 100 KDIYVVFELMESDLHQVIKANDDLTREHHQFFLYQMLRALKFMHTANVYHRDLKPKNIL 158
Score = 33.5 bits (75), Expect = 0.19, Method: Composition-based stats.
Identities = 25/80 (31%), Positives = 41/80 (51%), Gaps = 12/80 (15%)
Query 34 RAQILEAIHKQYIIYQLLRAIKYMHSGELLHRDMKPSNILLKKIFEAFHNATDAQRTFRE 93
R +ILE I K Y ++ A H+GE + +KKI + F + +DA R RE
Sbjct 24 RYRILEVIGKGS--YGVVCAAIDTHTGE---------KVAIKKINDVFEHISDALRILRE 72
Query 94 IMFLQQLAGHENIVRLMNVL 113
+ L+ L H +IV + +++
Sbjct 73 VKLLRLLR-HPDIVEIKSIM 91
> ath:AT5G19010 MPK16; MPK16; MAP kinase
Length=567
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 33/59 (55%), Positives = 44/59 (74%), Gaps = 1/59 (1%)
Query 16 RDIYLVFDFMETDLHAVIRAQI-LEAIHKQYIIYQLLRAIKYMHSGELLHRDMKPSNIL 73
RDIY+VF+ ME+DLH VI+A L H Q+ +YQLLR +KY+H+ + HRD+KP NIL
Sbjct 100 RDIYVVFELMESDLHQVIKANDDLTPEHYQFFLYQLLRGLKYIHTANVFHRDLKPKNIL 158
Score = 31.6 bits (70), Expect = 0.58, Method: Composition-based stats.
Identities = 19/56 (33%), Positives = 30/56 (53%), Gaps = 10/56 (17%)
Query 58 HSGELLHRDMKPSNILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
H+GE + +KKI + F + +DA R REI L+ L H +IV + ++L
Sbjct 46 HTGE---------KVAIKKINDIFEHVSDATRILREIKLLRLLR-HPDIVEIKHIL 91
> xla:397785 mapk1-b, Xp42, erk, erk2, mapk, mapk1b, mpk1; mitogen-activated
protein kinase 1 (EC:2.7.11.24); K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=361
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 35/77 (45%), Positives = 56/77 (72%), Gaps = 3/77 (3%)
Query 1 ENIVRLMNVLKA---DNDRDIYLVFDFMETDLHAVIRAQILEAIHKQYIIYQLLRAIKYM 57
ENI+ + ++++A + +D+Y+V D METDL+ +++ Q L H Y +YQ+LR +KY+
Sbjct 84 ENIIGINDIIRAPTIEQMKDVYIVQDLMETDLYKLLKTQHLSNDHICYFLYQILRGLKYI 143
Query 58 HSGELLHRDMKPSNILL 74
HS +LHRD+KPSN+LL
Sbjct 144 HSANVLHRDLKPSNLLL 160
> xla:398985 mapk1-a, MGC68934, erk, erk2, mapk, mapk1, mapk1a,
mpk1, xp42; mitogen-activated protein kinase 1 (EC:2.7.11.24);
K04371 extracellular signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=361
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 35/77 (45%), Positives = 56/77 (72%), Gaps = 3/77 (3%)
Query 1 ENIVRLMNVLKA---DNDRDIYLVFDFMETDLHAVIRAQILEAIHKQYIIYQLLRAIKYM 57
ENI+ + ++++A + +D+Y+V D METDL+ +++ Q L H Y +YQ+LR +KY+
Sbjct 84 ENIIGINDIIRAPTIEQMKDVYIVQDLMETDLYKLLKTQHLSNDHICYFLYQILRGLKYI 143
Query 58 HSGELLHRDMKPSNILL 74
HS +LHRD+KPSN+LL
Sbjct 144 HSANVLHRDLKPSNLLL 160
> ath:AT3G45640 ATMPK3; ATMPK3 (ARABIDOPSIS THALIANA MITOGEN-ACTIVATED
PROTEIN KINASE 3); MAP kinase/ kinase/ protein binding
/ protein kinase; K04371 extracellular signal-regulated
kinase 1/2 [EC:2.7.11.24]
Length=370
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 37/78 (47%), Positives = 54/78 (69%), Gaps = 4/78 (5%)
Query 1 ENIVRLMNVLKADNDR---DIYLVFDFMETDLHAVIRA-QILEAIHKQYIIYQLLRAIKY 56
ENI+ + +V+ R D+Y+ + M+TDLH +IR+ Q L H QY +YQLLR +KY
Sbjct 95 ENIIAIRDVVPPPLRRQFSDVYISTELMDTDLHQIIRSNQSLSEEHCQYFLYQLLRGLKY 154
Query 57 MHSGELLHRDMKPSNILL 74
+HS ++HRD+KPSN+LL
Sbjct 155 IHSANIIHRDLKPSNLLL 172
Score = 37.7 bits (86), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 20/42 (47%), Positives = 28/42 (66%), Gaps = 1/42 (2%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+ +KKI AF N DA+RT REI L+ L HENI+ + +V+
Sbjct 64 VAMKKIANAFDNHMDAKRTLREIKLLRHL-DHENIIAIRDVV 104
> mmu:26413 Mapk1, 9030612K14Rik, AA407128, AU018647, C78273,
ERK, Erk2, MAPK2, PRKM2, Prkm1, p41mapk, p42mapk; mitogen-activated
protein kinase 1 (EC:2.7.11.24); K04371 extracellular
signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=358
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 35/77 (45%), Positives = 56/77 (72%), Gaps = 3/77 (3%)
Query 1 ENIVRLMNVLKA---DNDRDIYLVFDFMETDLHAVIRAQILEAIHKQYIIYQLLRAIKYM 57
ENI+ + ++++A + +D+Y+V D METDL+ +++ Q L H Y +YQ+LR +KY+
Sbjct 79 ENIIGINDIIRAPTIEQMKDVYIVQDLMETDLYKLLKTQHLSNDHICYFLYQILRGLKYI 138
Query 58 HSGELLHRDMKPSNILL 74
HS +LHRD+KPSN+LL
Sbjct 139 HSANVLHRDLKPSNLLL 155
> hsa:5594 MAPK1, ERK, ERK2, ERT1, MAPK2, P42MAPK, PRKM1, PRKM2,
p38, p40, p41, p41mapk; mitogen-activated protein kinase
1 (EC:2.7.11.24); K04371 extracellular signal-regulated kinase
1/2 [EC:2.7.11.24]
Length=360
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 35/77 (45%), Positives = 56/77 (72%), Gaps = 3/77 (3%)
Query 1 ENIVRLMNVLKA---DNDRDIYLVFDFMETDLHAVIRAQILEAIHKQYIIYQLLRAIKYM 57
ENI+ + ++++A + +D+Y+V D METDL+ +++ Q L H Y +YQ+LR +KY+
Sbjct 81 ENIIGINDIIRAPTIEQMKDVYIVQDLMETDLYKLLKTQHLSNDHICYFLYQILRGLKYI 140
Query 58 HSGELLHRDMKPSNILL 74
HS +LHRD+KPSN+LL
Sbjct 141 HSANVLHRDLKPSNLLL 157
> ath:AT1G18150 ATMPK8; ATMPK8; MAP kinase
Length=589
Score = 75.1 bits (183), Expect = 5e-14, Method: Composition-based stats.
Identities = 33/59 (55%), Positives = 44/59 (74%), Gaps = 1/59 (1%)
Query 16 RDIYLVFDFMETDLHAVIRAQI-LEAIHKQYIIYQLLRAIKYMHSGELLHRDMKPSNIL 73
RDIY+VF+ ME+DLH VI+A L H Q+ +YQLLR +KY+H+ + HRD+KP NIL
Sbjct 179 RDIYVVFELMESDLHQVIKANDDLTPEHYQFFLYQLLRGLKYVHAANVFHRDLKPKNIL 237
Score = 32.0 bits (71), Expect = 0.53, Method: Composition-based stats.
Identities = 23/80 (28%), Positives = 40/80 (50%), Gaps = 12/80 (15%)
Query 34 RAQILEAIHKQYIIYQLLRAIKYMHSGELLHRDMKPSNILLKKIFEAFHNATDAQRTFRE 93
R QI E + K Y ++ + H+GE + +KKI + F + +DA R RE
Sbjct 103 RYQIQEVVGKGS--YGVVASAVDSHTGE---------RVAIKKINDVFEHVSDATRILRE 151
Query 94 IMFLQQLAGHENIVRLMNVL 113
I L+ L H ++V + +++
Sbjct 152 IKLLRLLR-HPDVVEIKHIM 170
> dre:541323 mapk7, bmk1, erk5, wu:fb73b02, zgc:113111; mitogen-activated
protein kinase 7 (EC:2.7.1.-); K04464 mitogen-activated
protein kinase 7 [EC:2.7.11.24]
Length=862
Score = 74.7 bits (182), Expect = 6e-14, Method: Composition-based stats.
Identities = 33/81 (40%), Positives = 55/81 (67%), Gaps = 5/81 (6%)
Query 1 ENIVRLMNVLKA----DNDRDIYLVFDFMETDLHAVIRA-QILEAIHKQYIIYQLLRAIK 55
+NI+ + ++L+ + +Y+V D ME+DLH +I + Q L H +Y +YQLLR +K
Sbjct 137 DNIIAIKDILQPVVPHSAFKSVYVVLDLMESDLHQIIHSRQPLTPEHTRYFLYQLLRGLK 196
Query 56 YMHSGELLHRDMKPSNILLKK 76
Y+HS ++HRD+KPSN+L+ +
Sbjct 197 YIHSANVIHRDLKPSNLLVNE 217
Score = 35.0 bits (79), Expect = 0.062, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 27/42 (64%), Gaps = 1/42 (2%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+ +KKI AF T+A+RT RE+ L+ H+NI+ + ++L
Sbjct 106 VAIKKIPNAFEVVTNAKRTLRELKILKHFK-HDNIIAIKDIL 146
> cpv:cgd3_3030 MAPK ; K04371 extracellular signal-regulated kinase
1/2 [EC:2.7.11.24]
Length=566
Score = 74.3 bits (181), Expect = 9e-14, Method: Composition-based stats.
Identities = 38/77 (49%), Positives = 53/77 (68%), Gaps = 3/77 (3%)
Query 1 ENIVRLMNVLKAD--NDRDIYLVFDFMETDLHAVIRA-QILEAIHKQYIIYQLLRAIKYM 57
ENI+ ++++L D N DIY+V METDLH VI + Q L H QY +YQ+LR + Y+
Sbjct 80 ENILGIIDLLPPDSPNFEDIYIVTQLMETDLHRVIYSKQTLTNEHIQYFMYQILRGLSYL 139
Query 58 HSGELLHRDMKPSNILL 74
H ++HRD+KPSNIL+
Sbjct 140 HKVNIIHRDLKPSNILV 156
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 22/44 (50%), Positives = 32/44 (72%), Gaps = 1/44 (2%)
Query 70 SNILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
S I +KKIF+AF + DA+R REI L+QL HENI+ ++++L
Sbjct 47 SYIAVKKIFDAFQDLIDAKRILREIKLLRQLQ-HENILGIIDLL 89
> ath:AT1G73670 ATMPK15; MAP kinase/ kinase
Length=576
Score = 74.3 bits (181), Expect = 9e-14, Method: Composition-based stats.
Identities = 32/59 (54%), Positives = 44/59 (74%), Gaps = 1/59 (1%)
Query 16 RDIYLVFDFMETDLHAVIRAQI-LEAIHKQYIIYQLLRAIKYMHSGELLHRDMKPSNIL 73
RD+Y+VF+ ME+DLH VI+A L H Q+ +YQLLR +KY+H+ + HRD+KP NIL
Sbjct 165 RDVYVVFELMESDLHQVIKANDDLTPEHHQFFLYQLLRGLKYVHAANVFHRDLKPKNIL 223
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 23/80 (28%), Positives = 40/80 (50%), Gaps = 12/80 (15%)
Query 34 RAQILEAIHKQYIIYQLLRAIKYMHSGELLHRDMKPSNILLKKIFEAFHNATDAQRTFRE 93
R QI E + K Y ++ + H+GE + +KKI + F + +DA R RE
Sbjct 89 RYQIQEVVGKGS--YGVVGSAIDTHTGE---------RVAIKKINDVFDHISDATRILRE 137
Query 94 IMFLQQLAGHENIVRLMNVL 113
I L+ L H ++V + +++
Sbjct 138 IKLLRLLL-HPDVVEIKHIM 156
> ath:AT3G18040 MPK9; MPK9 (MAP KINASE 9); MAP kinase
Length=510
Score = 73.6 bits (179), Expect = 1e-13, Method: Composition-based stats.
Identities = 32/59 (54%), Positives = 44/59 (74%), Gaps = 1/59 (1%)
Query 16 RDIYLVFDFMETDLHAVIRAQI-LEAIHKQYIIYQLLRAIKYMHSGELLHRDMKPSNIL 73
RDIY+VF+ ME+DLH VI+A L H Q+ +YQLLR +K++H+ + HRD+KP NIL
Sbjct 98 RDIYVVFELMESDLHQVIKANDDLTPEHYQFFLYQLLRGLKFIHTANVFHRDLKPKNIL 156
Score = 34.7 bits (78), Expect = 0.072, Method: Composition-based stats.
Identities = 31/101 (30%), Positives = 48/101 (47%), Gaps = 13/101 (12%)
Query 13 DNDRDIYLVFDFMETDLHAVIRAQILEAIHKQYIIYQLLRAIKYMHSGELLHRDMKPSNI 72
D + + L +F T+ R QI E I K Y ++ + HSGE +
Sbjct 2 DPHKKVALETEFF-TEYGEASRYQIQEVIGKGS--YGVVASAIDTHSGE---------KV 49
Query 73 LLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+KKI + F + +DA R REI L+ L H +IV + +V+
Sbjct 50 AIKKINDVFEHVSDATRILREIKLLRLLR-HPDIVEIKHVM 89
> cel:F43C1.2 mpk-1; MAP Kinase family member (mpk-1); K04371
extracellular signal-regulated kinase 1/2 [EC:2.7.11.24]
Length=444
Score = 73.2 bits (178), Expect = 2e-13, Method: Composition-based stats.
Identities = 32/62 (51%), Positives = 46/62 (74%), Gaps = 0/62 (0%)
Query 13 DNDRDIYLVFDFMETDLHAVIRAQILEAIHKQYIIYQLLRAIKYMHSGELLHRDMKPSNI 72
D+ +DIY+V METDL+ +++ Q L H Y +YQ+LR +KY+HS +LHRD+KPSN+
Sbjct 167 DSLKDIYIVQCLMETDLYKLLKTQKLSNDHVCYFLYQILRGLKYIHSANVLHRDLKPSNL 226
Query 73 LL 74
LL
Sbjct 227 LL 228
> ath:AT2G01450 ATMPK17; MAP kinase; K00924 [EC:2.7.1.-]
Length=486
Score = 73.2 bits (178), Expect = 2e-13, Method: Composition-based stats.
Identities = 31/59 (52%), Positives = 43/59 (72%), Gaps = 1/59 (1%)
Query 16 RDIYLVFDFMETDLHAVIRAQI-LEAIHKQYIIYQLLRAIKYMHSGELLHRDMKPSNIL 73
+DIY+VF+ ME+DLH V++ L H Q+ +YQLLR +K+MHS + HRD+KP NIL
Sbjct 91 KDIYVVFELMESDLHHVLKVNDDLTPQHHQFFLYQLLRGLKFMHSAHVFHRDLKPKNIL 149
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 22/72 (30%), Positives = 37/72 (51%), Gaps = 6/72 (8%)
Query 44 QYIIYQLLRAIKY--MHSGELLHRDMKPSNILLKKIFEAFHNATDAQRTFREIMFLQQLA 101
QY I +++ Y + S E H K + +KK+ F + +DA R REI L+ L
Sbjct 15 QYQIQEVVGKGSYGVVASAECPHTGGK---VAIKKMTNVFEHVSDAIRILREIKLLRLLR 71
Query 102 GHENIVRLMNVL 113
H +IV + +++
Sbjct 72 -HPDIVEIKHIM 82
> ath:AT2G46070 MPK12; MPK12 (MITOGEN-ACTIVATED PROTEIN KINASE
12); MAP kinase/ kinase; K04371 extracellular signal-regulated
kinase 1/2 [EC:2.7.11.24]
Length=372
Score = 72.4 bits (176), Expect = 4e-13, Method: Composition-based stats.
Identities = 31/78 (39%), Positives = 57/78 (73%), Gaps = 4/78 (5%)
Query 1 ENIVRLMNVLKA---DNDRDIYLVFDFMETDLHAVIRA-QILEAIHKQYIIYQLLRAIKY 56
EN++ + ++++ D D+Y+V++ M+TDL ++R+ Q L + ++++YQLLR +KY
Sbjct 98 ENVITIKDIVRPPQRDIFNDVYIVYELMDTDLQRILRSNQTLTSDQCRFLVYQLLRGLKY 157
Query 57 MHSGELLHRDMKPSNILL 74
+HS +LHRD++PSN+LL
Sbjct 158 VHSANILHRDLRPSNVLL 175
Score = 37.4 bits (85), Expect = 0.010, Method: Composition-based stats.
Identities = 17/42 (40%), Positives = 28/42 (66%), Gaps = 1/42 (2%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+ +KKI AF N DA+RT REI L+ + HEN++ + +++
Sbjct 67 VAIKKIGNAFDNIIDAKRTLREIKLLRHM-DHENVITIKDIV 107
> dre:266756 mapk14b, MAPK14, p38b, zp38b; mitogen-activated protein
kinase 14b (EC:2.7.11.24); K04441 p38 MAP kinase [EC:2.7.11.24]
Length=348
Score = 71.6 bits (174), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 33/83 (39%), Positives = 54/83 (65%), Gaps = 4/83 (4%)
Query 1 ENIVRLMNVLKADND----RDIYLVFDFMETDLHAVIRAQILEAIHKQYIIYQLLRAIKY 56
EN++ L++V D+YLV M DL+ +++ Q L H Q++IYQ+LRA+KY
Sbjct 82 ENVIGLLDVFSPATSLEGFNDVYLVTHLMGADLNNIVKCQKLTDDHVQFLIYQILRALKY 141
Query 57 MHSGELLHRDMKPSNILLKKIFE 79
+HS +++HRD+KPSN+ + + E
Sbjct 142 IHSADIIHRDLKPSNLAVNEDCE 164
Score = 32.0 bits (71), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 27/42 (64%), Gaps = 1/42 (2%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+ +KK+ F + A+RT+RE+ L+ + HEN++ L++V
Sbjct 51 VAVKKLSRPFQSIIHAKRTYRELRLLKHMK-HENVIGLLDVF 91
> ath:AT4G36450 ATMPK14 (Mitogen-activated protein kinase 14);
MAP kinase/ kinase; K04371 extracellular signal-regulated kinase
1/2 [EC:2.7.11.24]
Length=361
Score = 71.6 bits (174), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 33/78 (42%), Positives = 56/78 (71%), Gaps = 4/78 (5%)
Query 1 ENIVRLMNVLKADND---RDIYLVFDFMETDLHAVIRA-QILEAIHKQYIIYQLLRAIKY 56
EN++ L +V+ + RD+YLV++ M++DL+ +I++ Q L H +Y ++QLLR +KY
Sbjct 89 ENVISLKDVMLPTHRYSFRDVYLVYELMDSDLNQIIKSSQSLSDDHCKYFLFQLLRGLKY 148
Query 57 MHSGELLHRDMKPSNILL 74
+HS +LHRD+KP N+L+
Sbjct 149 LHSANILHRDLKPGNLLV 166
Score = 33.9 bits (76), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 26/42 (61%), Gaps = 1/42 (2%)
Query 72 ILLKKIFEAFHNATDAQRTFREIMFLQQLAGHENIVRLMNVL 113
+ +KKI F N DA RT RE+ L+ + HEN++ L +V+
Sbjct 58 VAIKKIHNVFENRIDALRTLRELKLLRHVR-HENVISLKDVM 98
> ath:AT2G42880 ATMPK20; MAP kinase
Length=606
Score = 71.6 bits (174), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 33/59 (55%), Positives = 45/59 (76%), Gaps = 1/59 (1%)
Query 16 RDIYLVFDFMETDLHAVIRAQI-LEAIHKQYIIYQLLRAIKYMHSGELLHRDMKPSNIL 73
+DIY+VF+ ME+DLH VI+A L H Q+ +YQLLRA+KY+H+ + HRD+KP NIL
Sbjct 100 KDIYVVFELMESDLHQVIKANDDLTREHYQFFLYQLLRALKYIHTANVYHRDLKPKNIL 158
> xla:100329215 p38-beta; mitogen-activated protein kinase p38
beta; K04441 p38 MAP kinase [EC:2.7.11.24]
Length=361
Score = 71.2 bits (173), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 33/83 (39%), Positives = 55/83 (66%), Gaps = 4/83 (4%)
Query 1 ENIVRLMNVL----KADNDRDIYLVFDFMETDLHAVIRAQILEAIHKQYIIYQLLRAIKY 56
EN++ L++V +N ++YLV + M DL+ +++ Q L H Q++IYQLLR +KY
Sbjct 80 ENVIGLLDVFTPSTSGENFNEVYLVTNLMGADLNNIVKCQKLTDDHIQFLIYQLLRGLKY 139
Query 57 MHSGELLHRDMKPSNILLKKIFE 79
+HS ++HRD+KPSN+ + + E
Sbjct 140 IHSAGIIHRDLKPSNLAVNEDCE 162
Lambda K H
0.330 0.142 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2052546600
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40