bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6333_orf2
Length=90
Score E
Sequences producing significant alignments: (Bits) Value
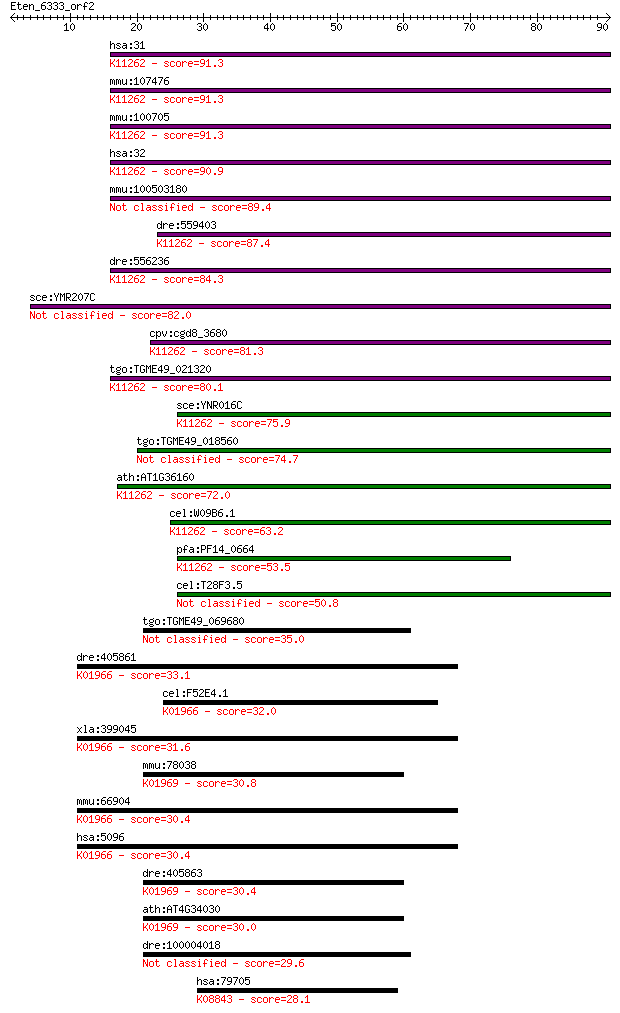
hsa:31 ACACA, ACAC, ACC, ACC1, ACCA; acetyl-CoA carboxylase al... 91.3 7e-19
mmu:107476 Acaca, A530025K05Rik, Acac, Acc1, Gm738; acetyl-Coe... 91.3 7e-19
mmu:100705 Acacb, AI597064, AW743042, Acc2, Accb; acetyl-Coenz... 91.3 7e-19
hsa:32 ACACB, ACC2, ACCB, HACC275; acetyl-CoA carboxylase beta... 90.9 1e-18
mmu:100503180 acetyl-CoA carboxylase 1-like 89.4 3e-18
dre:559403 acaca, im:7138837, si:ch211-199d18.1, wu:fj43d01; a... 87.4 9e-18
dre:556236 acetyl-CoA carboxylase 1-like; K11262 acetyl-CoA ca... 84.3 8e-17
sce:YMR207C HFA1; Hfa1p (EC:6.4.1.2 6.3.4.14) 82.0 4e-16
cpv:cgd8_3680 acetyl-CoA carboxylase like biotin dependent car... 81.3 8e-16
tgo:TGME49_021320 acetyl-CoA carboxylase, putative (EC:6.3.4.1... 80.1 1e-15
sce:YNR016C ACC1, ABP2, FAS3, MTR7; Acc1p (EC:6.4.1.2 6.3.4.14... 75.9 3e-14
tgo:TGME49_018560 acetyl-coA carboxylase (EC:6.3.4.14 2.3.1.12... 74.7 6e-14
ath:AT1G36160 ACC1; ACC1 (ACETYL-COENZYME A CARBOXYLASE 1); ac... 72.0 4e-13
cel:W09B6.1 pod-2; Polarity and Osmotic sensitivity Defect fam... 63.2 2e-10
pfa:PF14_0664 ACC1; biotin carboxylase subunit of acetyl CoA c... 53.5 2e-07
cel:T28F3.5 hypothetical protein 50.8 9e-07
tgo:TGME49_069680 acyl-CoA carboxyltransferase beta chain, put... 35.0 0.065
dre:405861 pccb, MGC85813, zgc:85813; propionyl Coenzyme A car... 33.1 0.23
cel:F52E4.1 pccb-1; Propionyl Coenzyme A Carboxylase Beta subu... 32.0 0.43
xla:399045 pccb, MGC68650; propionyl CoA carboxylase, beta pol... 31.6 0.59
mmu:78038 Mccc2, 4930552N12Rik, MCCB; methylcrotonoyl-Coenzyme... 30.8 0.99
mmu:66904 Pccb, 1300012P06Rik, AI314687, R74805; propionyl Coe... 30.4 1.2
hsa:5096 PCCB, DKFZp451E113; propionyl CoA carboxylase, beta p... 30.4 1.3
dre:405863 mccc2, MGC85685, si:dkey-57m14.1, zgc:85685; methyl... 30.4 1.4
ath:AT4G34030 MCCB; MCCB (3-METHYLCROTONYL-COA CARBOXYLASE); b... 30.0 2.1
dre:100004018 im:7143041; si:ch211-198n5.11 29.6 2.5
hsa:79705 LRRK1, FLJ23119, FLJ27465, KIAA1790, RIPK6, Roco1; l... 28.1 7.0
> hsa:31 ACACA, ACAC, ACC, ACC1, ACCA; acetyl-CoA carboxylase
alpha (EC:6.4.1.2 6.3.4.14); K11262 acetyl-CoA carboxylase /
biotin carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2383
Score = 91.3 bits (225), Expect = 7e-19, Method: Composition-based stats.
Identities = 43/75 (57%), Positives = 50/75 (66%), Gaps = 0/75 (0%)
Query 16 HKKHQRRWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPA 75
H + +W+SGF D GSF E+ PW VVVGRARLGG P+GV+AV+ RT PADPA
Sbjct 1980 HPTQKGQWLSGFFDYGSFSEIMQPWAQTVVVGRARLGGIPVGVVAVETRTVELSIPADPA 2039
Query 76 NADSARVEIPQAGQV 90
N DS I QAGQV
Sbjct 2040 NLDSEAKIIQQAGQV 2054
> mmu:107476 Acaca, A530025K05Rik, Acac, Acc1, Gm738; acetyl-Coenzyme
A carboxylase alpha (EC:6.4.1.2 6.3.4.14); K11262 acetyl-CoA
carboxylase / biotin carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2345
Score = 91.3 bits (225), Expect = 7e-19, Method: Composition-based stats.
Identities = 43/75 (57%), Positives = 50/75 (66%), Gaps = 0/75 (0%)
Query 16 HKKHQRRWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPA 75
H + +W+SGF D GSF E+ PW VVVGRARLGG P+GV+AV+ RT PADPA
Sbjct 1942 HPTQKGQWLSGFFDYGSFSEIMQPWAQTVVVGRARLGGIPVGVVAVETRTVELSIPADPA 2001
Query 76 NADSARVEIPQAGQV 90
N DS I QAGQV
Sbjct 2002 NLDSEAKIIQQAGQV 2016
> mmu:100705 Acacb, AI597064, AW743042, Acc2, Accb; acetyl-Coenzyme
A carboxylase beta (EC:6.4.1.2); K11262 acetyl-CoA carboxylase
/ biotin carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2448
Score = 91.3 bits (225), Expect = 7e-19, Method: Composition-based stats.
Identities = 44/75 (58%), Positives = 48/75 (64%), Gaps = 0/75 (0%)
Query 16 HKKHQRRWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPA 75
H + W SGF D GSFKE+ PW VV GRARLGG P+GVIAV+ RT PADPA
Sbjct 2044 HPTLKGTWQSGFFDHGSFKEIMAPWAQTVVTGRARLGGIPVGVIAVETRTVEVAVPADPA 2103
Query 76 NADSARVEIPQAGQV 90
N DS I QAGQV
Sbjct 2104 NLDSEAKIIQQAGQV 2118
> hsa:32 ACACB, ACC2, ACCB, HACC275; acetyl-CoA carboxylase beta
(EC:6.4.1.2 6.3.4.14); K11262 acetyl-CoA carboxylase / biotin
carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2458
Score = 90.9 bits (224), Expect = 1e-18, Method: Composition-based stats.
Identities = 44/75 (58%), Positives = 48/75 (64%), Gaps = 0/75 (0%)
Query 16 HKKHQRRWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPA 75
H + W SGF D GSFKE+ PW VV GRARLGG P+GVIAV+ RT PADPA
Sbjct 2054 HPTLKGTWQSGFFDHGSFKEIMAPWAQTVVTGRARLGGIPVGVIAVETRTVEVAVPADPA 2113
Query 76 NADSARVEIPQAGQV 90
N DS I QAGQV
Sbjct 2114 NLDSEAKIIQQAGQV 2128
> mmu:100503180 acetyl-CoA carboxylase 1-like
Length=580
Score = 89.4 bits (220), Expect = 3e-18, Method: Composition-based stats.
Identities = 43/75 (57%), Positives = 50/75 (66%), Gaps = 0/75 (0%)
Query 16 HKKHQRRWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPA 75
H + +W+SGF D GSF E+ PW VVVGRARLGG P+GV+AV+ RT PADPA
Sbjct 197 HPTQKGQWLSGFFDYGSFSEIMQPWAQTVVVGRARLGGIPVGVVAVETRTVELSIPADPA 256
Query 76 NADSARVEIPQAGQV 90
N DS I QAGQV
Sbjct 257 NLDSEAKIIQQAGQV 271
> dre:559403 acaca, im:7138837, si:ch211-199d18.1, wu:fj43d01;
acetyl-Coenzyme A carboxylase alpha; K11262 acetyl-CoA carboxylase
/ biotin carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2349
Score = 87.4 bits (215), Expect = 9e-18, Method: Composition-based stats.
Identities = 42/68 (61%), Positives = 47/68 (69%), Gaps = 0/68 (0%)
Query 23 WVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPANADSARV 82
WVSGF D+GSF E+ PW VVVGRARLGG P GV+AV+ R+ PADPAN DS
Sbjct 1952 WVSGFFDQGSFLEIMQPWAQSVVVGRARLGGIPTGVVAVETRSVELSIPADPANLDSEAK 2011
Query 83 EIPQAGQV 90
I QAGQV
Sbjct 2012 IIQQAGQV 2019
> dre:556236 acetyl-CoA carboxylase 1-like; K11262 acetyl-CoA
carboxylase / biotin carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2240
Score = 84.3 bits (207), Expect = 8e-17, Method: Composition-based stats.
Identities = 42/75 (56%), Positives = 47/75 (62%), Gaps = 0/75 (0%)
Query 16 HKKHQRRWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPA 75
H + W SGF D GSF E+ W VVVGRARLGG P+GVIAV+ RT PADPA
Sbjct 1836 HPTVKGAWQSGFFDHGSFLEIMATWAQTVVVGRARLGGIPLGVIAVETRTVELAIPADPA 1895
Query 76 NADSARVEIPQAGQV 90
N DS + QAGQV
Sbjct 1896 NLDSEAKLLQQAGQV 1910
> sce:YMR207C HFA1; Hfa1p (EC:6.4.1.2 6.3.4.14)
Length=2123
Score = 82.0 bits (201), Expect = 4e-16, Method: Composition-based stats.
Identities = 41/93 (44%), Positives = 55/93 (59%), Gaps = 6/93 (6%)
Query 4 DAEGAAEVPQQQHKKHQRRW------VSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIG 57
D + A +VP + + +W SG D+ SF E W GV+VGRARLGG P+G
Sbjct 1727 DFKPAKQVPYEARWLIEGKWDSNNNFQSGLFDKDSFFETLSGWAKGVIVGRARLGGIPVG 1786
Query 58 VIAVDPRTTTAVAPADPANADSARVEIPQAGQV 90
VIAV+ +T + PADPAN DS+ + +AGQV
Sbjct 1787 VIAVETKTIEEIIPADPANLDSSEFSVKEAGQV 1819
> cpv:cgd8_3680 acetyl-CoA carboxylase like biotin dependent carboxylase
involved in fatty acid biosynthesis ; K11262 acetyl-CoA
carboxylase / biotin carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2733
Score = 81.3 bits (199), Expect = 8e-16, Method: Composition-based stats.
Identities = 32/69 (46%), Positives = 45/69 (65%), Gaps = 0/69 (0%)
Query 22 RWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPANADSAR 81
RW+SG CD+ SFKEV W V+VGR R+GG P+G I V+ R T + PADP ++
Sbjct 2389 RWLSGLCDKDSFKEVMSDWAKSVIVGRGRIGGIPVGFILVETRVTEYIQPADPVMPHTSE 2448
Query 82 VEIPQAGQV 90
+ + +AGQ+
Sbjct 2449 LRVTRAGQI 2457
> tgo:TGME49_021320 acetyl-CoA carboxylase, putative (EC:6.3.4.14
2.3.1.12 6.4.1.3); K11262 acetyl-CoA carboxylase / biotin
carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2599
Score = 80.1 bits (196), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 41/75 (54%), Positives = 48/75 (64%), Gaps = 0/75 (0%)
Query 16 HKKHQRRWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPA 75
+ K +VSGF D+ SFKE WG VVVGRARLGG P G IAV+ RTT A PADP+
Sbjct 2237 YTKEDGTFVSGFFDKNSFKEYLAGWGKSVVVGRARLGGIPFGAIAVETRTTEARVPADPS 2296
Query 76 NADSARVEIPQAGQV 90
+ DS + AGQV
Sbjct 2297 SPDSRESVVMHAGQV 2311
> sce:YNR016C ACC1, ABP2, FAS3, MTR7; Acc1p (EC:6.4.1.2 6.3.4.14);
K11262 acetyl-CoA carboxylase / biotin carboxylase [EC:6.4.1.2
6.3.4.14]
Length=2233
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 37/65 (56%), Positives = 43/65 (66%), Gaps = 0/65 (0%)
Query 26 GFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPANADSARVEIP 85
G D+GSF E W GVVVGRARLGG P+GVI V+ RT + PADPAN +SA I
Sbjct 1859 GLFDKGSFFETLSGWAKGVVVGRARLGGIPLGVIGVETRTVENLIPADPANPNSAETLIQ 1918
Query 86 QAGQV 90
+ GQV
Sbjct 1919 EPGQV 1923
> tgo:TGME49_018560 acetyl-coA carboxylase (EC:6.3.4.14 2.3.1.12
6.4.1.3 3.2.1.17)
Length=3399
Score = 74.7 bits (182), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 34/71 (47%), Positives = 47/71 (66%), Gaps = 0/71 (0%)
Query 20 QRRWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPANADS 79
Q W+ G DRGS++E W V++GRARLGG P+GV+AV+ R T A PADPA +
Sbjct 2974 QGNWLGGVFDRGSYREAMADWARSVIIGRARLGGIPVGVVAVETRVTEAKQPADPAMPHT 3033
Query 80 ARVEIPQAGQV 90
+ + + +AGQV
Sbjct 3034 SEILLTRAGQV 3044
> ath:AT1G36160 ACC1; ACC1 (ACETYL-COENZYME A CARBOXYLASE 1);
acetyl-CoA carboxylase; K11262 acetyl-CoA carboxylase / biotin
carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2254
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 34/74 (45%), Positives = 45/74 (60%), Gaps = 0/74 (0%)
Query 17 KKHQRRWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPAN 76
K + +W+ G D+ SF E W VV GRA+LGG P+GV+AV+ +T + PADP
Sbjct 1858 KDNTGKWLGGIFDKNSFIETLEGWARTVVTGRAKLGGIPVGVVAVETQTVMQIIPADPGQ 1917
Query 77 ADSARVEIPQAGQV 90
DS +PQAGQV
Sbjct 1918 LDSHERVVPQAGQV 1931
> cel:W09B6.1 pod-2; Polarity and Osmotic sensitivity Defect family
member (pod-2); K11262 acetyl-CoA carboxylase / biotin
carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2054
Score = 63.2 bits (152), Expect = 2e-10, Method: Composition-based stats.
Identities = 30/66 (45%), Positives = 39/66 (59%), Gaps = 0/66 (0%)
Query 25 SGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPANADSARVEI 84
+G CD SF E+ G W +V GRARL G PIGV++ + R + + PADPA S
Sbjct 1705 TGICDTMSFDEICGDWAKSIVAGRARLCGIPIGVVSSEFRNFSTIVPADPAIDGSQVQNT 1764
Query 85 PQAGQV 90
+AGQV
Sbjct 1765 QRAGQV 1770
> pfa:PF14_0664 ACC1; biotin carboxylase subunit of acetyl CoA
carboxylase, putative (EC:6.4.1.2); K11262 acetyl-CoA carboxylase
/ biotin carboxylase [EC:6.4.1.2 6.3.4.14]
Length=3367
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 21/50 (42%), Positives = 29/50 (58%), Gaps = 0/50 (0%)
Query 26 GFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPA 75
GF D+ ++ E WG G++ GR +LG P+G IAV+ T P DPA
Sbjct 3010 GFLDKNTYFEYMNEWGKGIITGRGKLGSIPVGFIAVNKNLVTQSIPCDPA 3059
> cel:T28F3.5 hypothetical protein
Length=1679
Score = 50.8 bits (120), Expect = 9e-07, Method: Composition-based stats.
Identities = 25/65 (38%), Positives = 37/65 (56%), Gaps = 0/65 (0%)
Query 26 GFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTTAVAPADPANADSARVEIP 85
G D SF E++ W A +V GRA+L G P+GVIA ++ + AD + +S +
Sbjct 1385 GLFDTDSFDEIRSSWAASMVTGRAKLNGLPVGVIASQWKSYEKLLLADESVENSEEMITS 1444
Query 86 QAGQV 90
+AGQV
Sbjct 1445 KAGQV 1449
> tgo:TGME49_069680 acyl-CoA carboxyltransferase beta chain, putative
(EC:6.4.1.3)
Length=632
Score = 35.0 bits (79), Expect = 0.065, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 21 RRWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIA 60
+R ++ DR + E + +G +V G A L G PIGV+A
Sbjct 371 KRLLACLLDRSALAEFKPLYGETLVCGFAHLNGYPIGVMA 410
> dre:405861 pccb, MGC85813, zgc:85813; propionyl Coenzyme A carboxylase,
beta polypeptide (EC:6.4.1.3); K01966 propionyl-CoA
carboxylase beta chain [EC:6.4.1.3]
Length=557
Score = 33.1 bits (74), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 16/58 (27%), Positives = 28/58 (48%), Gaps = 1/58 (1%)
Query 11 VPQQQHKKHQRR-WVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTT 67
VP + K + V G D F E+ + +VVG AR+ G+ +G++ P+ +
Sbjct 322 VPFESTKAYDMLDIVHGIVDEREFFEIMPNYAKNIVVGFARMNGRTVGIVGNQPKVAS 379
> cel:F52E4.1 pccb-1; Propionyl Coenzyme A Carboxylase Beta subunit
family member (pccb-1); K01966 propionyl-CoA carboxylase
beta chain [EC:6.4.1.3]
Length=522
Score = 32.0 bits (71), Expect = 0.43, Method: Composition-based stats.
Identities = 12/41 (29%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 24 VSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPR 64
V D G F E+ + +V+G AR+ G+ +G++ +P+
Sbjct 302 VHALVDEGDFFEIMPDYAKNLVIGFARMNGRTVGIVGNNPK 342
> xla:399045 pccb, MGC68650; propionyl CoA carboxylase, beta polypeptide
(EC:6.4.1.3); K01966 propionyl-CoA carboxylase beta
chain [EC:6.4.1.3]
Length=541
Score = 31.6 bits (70), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 14/58 (24%), Positives = 27/58 (46%), Gaps = 1/58 (1%)
Query 11 VPQQQHKKHQRR-WVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTT 67
VP + K + + D F E+ + +VVG AR+ G+ +G++ P+ +
Sbjct 308 VPMESTKAYDMLDIIHSIIDEREFFEIMPNYAKNIVVGFARMNGRTVGIVGNQPKVAS 365
> mmu:78038 Mccc2, 4930552N12Rik, MCCB; methylcrotonoyl-Coenzyme
A carboxylase 2 (beta) (EC:6.4.1.4); K01969 3-methylcrotonyl-CoA
carboxylase beta subunit [EC:6.4.1.4]
Length=563
Score = 30.8 bits (68), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 13/39 (33%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 21 RRWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVI 59
R ++ D F E + +G +V G AR+ G P+G+I
Sbjct 332 REVIARIVDGSRFNEFKALYGDTLVTGFARIFGYPVGII 370
> mmu:66904 Pccb, 1300012P06Rik, AI314687, R74805; propionyl Coenzyme
A carboxylase, beta polypeptide (EC:6.4.1.3); K01966
propionyl-CoA carboxylase beta chain [EC:6.4.1.3]
Length=541
Score = 30.4 bits (67), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 14/58 (24%), Positives = 26/58 (44%), Gaps = 1/58 (1%)
Query 11 VPQQQHKKHQRR-WVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTT 67
VP + K + + D F E+ + +VVG AR+ G+ +G++ P +
Sbjct 308 VPLESSKAYNMLDIIHAVIDEREFFEIMPSYAKNIVVGFARMNGRTVGIVGNQPNVAS 365
> hsa:5096 PCCB, DKFZp451E113; propionyl CoA carboxylase, beta
polypeptide (EC:6.4.1.3); K01966 propionyl-CoA carboxylase
beta chain [EC:6.4.1.3]
Length=539
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 13/58 (22%), Positives = 27/58 (46%), Gaps = 1/58 (1%)
Query 11 VPQQQHKKHQR-RWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIAVDPRTTT 67
VP + K + + D F E+ + ++VG AR+ G+ +G++ P+ +
Sbjct 306 VPLESTKAYNMVDIIHSVVDEREFFEIMPNYAKNIIVGFARMNGRTVGIVGNQPKVAS 363
> dre:405863 mccc2, MGC85685, si:dkey-57m14.1, zgc:85685; methylcrotonoyl-Coenzyme
A carboxylase 2 (beta) (EC:6.4.1.4); K01969
3-methylcrotonyl-CoA carboxylase beta subunit [EC:6.4.1.4]
Length=566
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 12/39 (30%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 21 RRWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVI 59
R ++ D F E + +G ++ G AR+ G P+G+I
Sbjct 335 REVIARIVDGSKFDEFKAFYGDTLITGFARIFGYPVGII 373
> ath:AT4G34030 MCCB; MCCB (3-METHYLCROTONYL-COA CARBOXYLASE);
biotin carboxylase/ methylcrotonoyl-CoA carboxylase (EC:6.4.1.4);
K01969 3-methylcrotonyl-CoA carboxylase beta subunit
[EC:6.4.1.4]
Length=587
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 13/39 (33%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 21 RRWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVI 59
R ++ D F E + +G +V G AR+ GQ +G+I
Sbjct 356 RSIIARIVDGSEFDEFKKQYGTTLVTGFARIYGQTVGII 394
> dre:100004018 im:7143041; si:ch211-198n5.11
Length=603
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 21 RRWVSGFCDRGSFKEVQGPWGAGVVVGRARLGGQPIGVIA 60
R VS D F+E + +G ++ G AR+ G IG++A
Sbjct 381 RLIVSRLTDGSRFQEFKARYGTTLLTGFARIDGHQIGIVA 420
> hsa:79705 LRRK1, FLJ23119, FLJ27465, KIAA1790, RIPK6, Roco1;
leucine-rich repeat kinase 1 (EC:2.7.11.1); K08843 leucine-rich
repeat kinase 1 [EC:2.7.11.1]
Length=2015
Score = 28.1 bits (61), Expect = 7.0, Method: Composition-based stats.
Identities = 15/30 (50%), Positives = 18/30 (60%), Gaps = 3/30 (10%)
Query 29 DRGSFKEVQGPWGAGVVVGRARLGGQPIGV 58
D GS V G G+G V+ RAR GQP+ V
Sbjct 1243 DEGS---VLGQGGSGTVIYRARYQGQPVAV 1269
Lambda K H
0.315 0.132 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2012433808
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40