bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
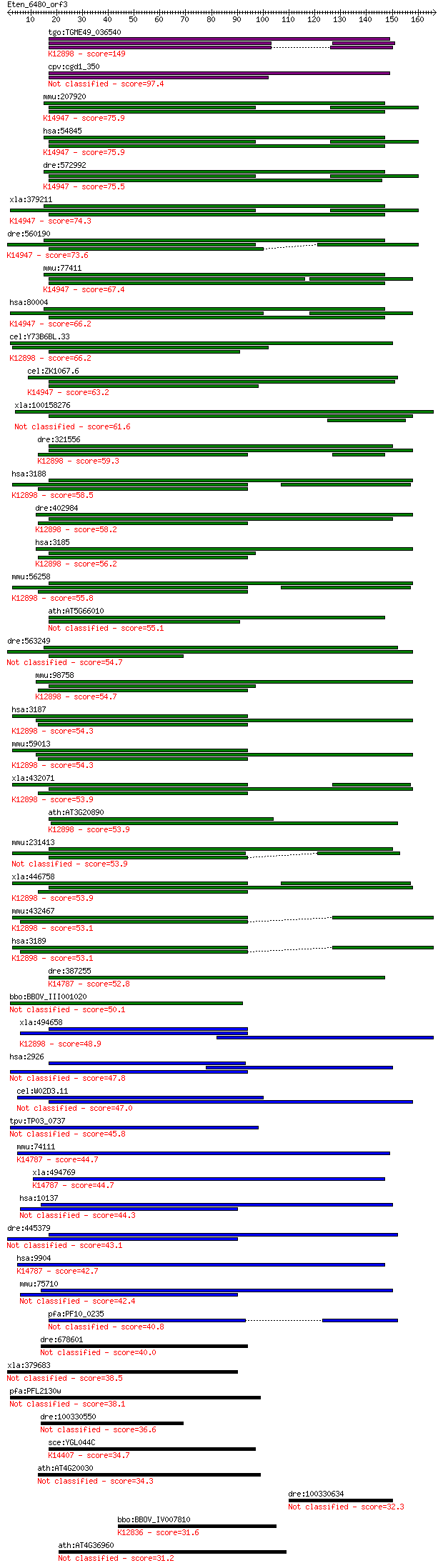
Query= Eten_6480_orf3
Length=166
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_036540 RRM domain-containing protein ; K12898 heter... 149 6e-36
cpv:cgd1_350 CG8205/fusilli (animal)- like, 2x RRM domains, in... 97.4 2e-20
mmu:207920 Esrp1, 2210008M09Rik, A630065D16, BC031468, MGC2580... 75.9 7e-14
hsa:54845 ESRP1, FLJ20171, RBM35A, RMB35A; epithelial splicing... 75.9 7e-14
dre:572992 esrp2, FLJ20171, MGC55721, MGC77254, cb404, fa07a06... 75.5 8e-14
xla:379211 esrp1, MGC53361, rbm35a; epithelial splicing regula... 74.3 2e-13
dre:560190 esrp1, rbm35a, wu:fi28a07, zgc:154050; epithelial s... 73.6 3e-13
mmu:77411 Esrp2, 9530027K23Rik, Rbm35b; epithelial splicing re... 67.4 2e-11
hsa:80004 ESRP2, FLJ21918, FLJ22248, RBM35B; epithelial splici... 66.2 4e-11
cel:Y73B6BL.33 hypothetical protein; K12898 heterogeneous nucl... 66.2 5e-11
cel:ZK1067.6 sym-2; SYnthetic lethal with Mec family member (s... 63.2 4e-10
xla:100158276 grsf1; G-rich RNA sequence binding factor 1 61.6
dre:321556 hnrnph1l, fb19a06, wu:fb19a06, zgc:55399, zgc:85960... 59.3 5e-09
hsa:3188 HNRNPH2, FTP3, HNRPH', HNRPH2, hnRNPH'; heterogeneous... 58.5 1e-08
dre:402984 hnrnph1, MGC77712, zgc:77712; heterogeneous nuclear... 58.2 1e-08
hsa:3185 HNRNPF, HNRPF, MGC110997, OK/SW-cl.23, mcs94-1; heter... 56.2 5e-08
mmu:56258 Hnrnph2, DXHXS1271E, Ftp-3, Ftp3, H', HNRNP, Hnrph2;... 55.8 7e-08
ath:AT5G66010 RNA binding / nucleic acid binding / nucleotide ... 55.1 1e-07
dre:563249 grsf1, MGC153305, wu:fb62c04, zgc:153305; G-rich RN... 54.7 1e-07
mmu:98758 Hnrnpf, 4833420I20Rik, AA407306, Hnrpf, MGC101981, M... 54.7 2e-07
hsa:3187 HNRNPH1, DKFZp686A15170, HNRPH, HNRPH1, hnRNPH; heter... 54.3 2e-07
mmu:59013 Hnrnph1, AI642080, E430005G16Rik, Hnrnph, Hnrph1; he... 54.3 2e-07
xla:432071 hnrnph1-a, MGC78776, hnrnph, hnrnph2, hnrph, hnrph1... 53.9 2e-07
ath:AT3G20890 RNA binding / nucleic acid binding / nucleotide ... 53.9 2e-07
mmu:231413 Grsf1, B130010H02, BB232551, C80280, D5Wsu31e; G-ri... 53.9 2e-07
xla:446758 hnrnph1-b, MGC130700, MGC80081, hnrnph, hnrnph1, hn... 53.9 2e-07
mmu:432467 Hnrnph3, AA693301, AI666703, Hnrph3; heterogeneous ... 53.1 4e-07
hsa:3189 HNRNPH3, 2H9, FLJ34092, HNRPH3; heterogeneous nuclear... 53.1 4e-07
dre:387255 rbm19, npo; RNA binding motif protein 19; K14787 mu... 52.8 5e-07
bbo:BBOV_III001020 17.m07117; hypothetical protein 50.1 3e-06
xla:494658 hnrnph3, hnrph3; heterogeneous nuclear ribonucleopr... 48.9 7e-06
hsa:2926 GRSF1, FLJ13125; G-rich RNA sequence binding factor 1 47.8
cel:W02D3.11 hrpf-1; HnRNP F homolog family member (hrpf-1) 47.0 3e-05
tpv:TP03_0737 hypothetical protein 45.8 6e-05
mmu:74111 Rbm19, 1200009A02Rik, AV302714, KIAA0682, NPO; RNA b... 44.7 1e-04
xla:494769 rbm19; RNA binding motif protein 19; K14787 multipl... 44.7 2e-04
hsa:10137 RBM12, HRIHFB2091, KIAA0765, SWAN; RNA binding motif... 44.3 2e-04
dre:445379 rbm12, zgc:193560, zgc:193570; RNA binding motif pr... 43.1 5e-04
hsa:9904 RBM19, DKFZp586F1023, KIAA0682; RNA binding motif pro... 42.7 6e-04
mmu:75710 Rbm12, 5730420G12Rik, 9430070C08Rik, AI852903, MGC30... 42.4 6e-04
pfa:PF10_0235 RNA binding protein, putative 40.8 0.002
dre:678601 MGC136953; zgc:136953 40.0 0.003
xla:379683 rbm12, MGC68861, SWAN; RNA binding motif protein 12 38.5
pfa:PFL2130w conserved Plasmodium protein 38.1 0.015
dre:100330550 heterogeneous nuclear ribonucleoprotein H3-like 36.6 0.035
sce:YGL044C RNA15; Cleavage and polyadenylation factor I (CF I... 34.7 0.17
ath:AT4G20030 RNA recognition motif (RRM)-containing protein 34.3 0.18
dre:100330634 RNA binding motif protein 12-like 32.3 0.85
bbo:BBOV_IV007810 23.m06352; U2 splicing factor subunit; K1283... 31.6 1.2
ath:AT4G36960 RNA recognition motif (RRM)-containing protein 31.2 1.7
> tgo:TGME49_036540 RRM domain-containing protein ; K12898 heterogeneous
nuclear ribonucleoprotein F/H
Length=513
Score = 149 bits (375), Expect = 6e-36, Method: Compositional matrix adjust.
Identities = 67/134 (50%), Positives = 97/134 (72%), Gaps = 3/134 (2%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVRM 76
+RLRGLPWDV E+ +I F KPVV + +V +C+G +KR TGEAYV + +R+ A++
Sbjct 54 VRLRGLPWDVQEENVIAFFKPVVALENDNVLICIGFDKRTTGEAYVQLPDPGLREQAIKD 113
Query 77 LHGRMMGARWIEVFRSSAEDFERAQQRRVAMLS--SDSRDGRDADVKVLNLTVLKLRGLP 134
LHGR++G RWIEVFR+S E+F++A RR +++ S + D DA + +NL V+KLRGLP
Sbjct 114 LHGRLLGTRWIEVFRASEEEFQKADDRRKTVMAAISGNTDSLDAS-RRMNLNVVKLRGLP 172
Query 135 WTCTEMNVVNFFKS 148
W+C+E +V FFK+
Sbjct 173 WSCSENEIVRFFKA 186
Score = 65.9 bits (159), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 32/88 (36%), Positives = 57/88 (64%), Gaps = 2/88 (2%)
Query 17 IRLRGLPWDVNEDAIIRFVKPV--VEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAV 74
++LRGLPW +E+ I+RF K +I DV + V + R++G A+V + + ++ + A
Sbjct 166 VKLRGLPWSCSENEIVRFFKAEGGFDIHSDDVVLGVTGDGRLSGIAFVELPSPDVAEKAR 225
Query 75 RMLHGRMMGARWIEVFRSSAEDFERAQQ 102
+LH + MG R+IEV+ ++ ED +RA++
Sbjct 226 EVLHKKYMGRRFIEVYPATREDMQRAKR 253
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/86 (32%), Positives = 43/86 (50%), Gaps = 2/86 (2%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVRM 76
+RLRGLP+ NE I++F + + + ++ R +GEAYV A +
Sbjct 416 LRLRGLPYSANEQHIVQFFHGF--HMAAILPSTIPIDGRPSGEAYVQFVDAAEALRAFQA 473
Query 77 LHGRMMGARWIEVFRSSAEDFERAQQ 102
+G M R IE+F SS ++ E A Q
Sbjct 474 KNGGRMDKRMIELFPSSKQEMEFAAQ 499
Score = 35.8 bits (81), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 14/24 (58%), Positives = 18/24 (75%), Gaps = 0/24 (0%)
Query 127 VLKLRGLPWTCTEMNVVNFFKSMV 150
V++LRGLPW E NV+ FFK +V
Sbjct 53 VVRLRGLPWDVQEENVIAFFKPVV 76
Score = 32.3 bits (72), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 11/24 (45%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 126 TVLKLRGLPWTCTEMNVVNFFKSM 149
VL+LRGLP++ E ++V FF
Sbjct 414 QVLRLRGLPYSANEQHIVQFFHGF 437
> cpv:cgd1_350 CG8205/fusilli (animal)- like, 2x RRM domains,
involved in RNA metabolism
Length=569
Score = 97.4 bits (241), Expect = 2e-20, Method: Composition-based stats.
Identities = 51/136 (37%), Positives = 78/136 (57%), Gaps = 5/136 (3%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVRM 76
+RLRGLPW II F P+ I D+ + + ++TGEAYV + + + ++++ +
Sbjct 318 VRLRGLPWKAAVLDIIAFFNPICRISSYDIAISYNKDGKMTGEAYVLLPSIKAYELSLTL 377
Query 77 LHGRMMGARWIEVFRSSAEDFERAQQ----RRVAMLSSDSRDGRDADVKVLNLTVLKLRG 132
LHG+ MG RWIEV SS ++F Q ++ S D + D + N +VL+LRG
Sbjct 378 LHGKRMGKRWIEVLPSSTKEFLICLQITSLKKQNQNPSIFNDNKIID-RYYNRSVLRLRG 436
Query 133 LPWTCTEMNVVNFFKS 148
LPW+ TE+ +V FF S
Sbjct 437 LPWSTTEIEIVQFFIS 452
Score = 56.6 bits (135), Expect = 3e-08, Method: Composition-based stats.
Identities = 33/88 (37%), Positives = 54/88 (61%), Gaps = 4/88 (4%)
Query 17 IRLRGLPWDVNEDAIIRFVKP--VVEIRESDVCVCVGLNKRVTGEAYVNV-HTTEMRDMA 73
+RLRGLPW E I++F + + SDV + + N+R +GEA++ + H + D A
Sbjct 432 LRLRGLPWSTTEIEIVQFFISGGIYGLNASDVFLGITENQRASGEAWIILPHKCDAFD-A 490
Query 74 VRMLHGRMMGARWIEVFRSSAEDFERAQ 101
R+L+ R++G R+IEVF SS ++ A+
Sbjct 491 QRILNRRVIGKRYIEVFISSFQELTTAR 518
> mmu:207920 Esrp1, 2210008M09Rik, A630065D16, BC031468, MGC25805,
Rbm35a; epithelial splicing regulatory protein 1; K14947
epithelial splicing regulatory protein 1/2
Length=681
Score = 75.9 bits (185), Expect = 7e-14, Method: Composition-based stats.
Identities = 46/135 (34%), Positives = 68/135 (50%), Gaps = 16/135 (11%)
Query 15 THIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAV 74
T +R RGLPW ++ I RF K + I + +C+ R GEA V + E RD+A+
Sbjct 225 TVVRARGLPWQSSDQDIARFFKGL-NIAKGGAALCLNAQGRRNGEALVRFVSEEHRDLAL 283
Query 75 RMLHGRMMGARWIEVFRSSAEDFER---AQQRRVAMLSSDSRDGRDADVKVLNLTVLKLR 131
+ H MG R+IEV++++ EDF + VA S N ++++R
Sbjct 284 QR-HKHHMGTRYIEVYKATGEDFLKIAGGTSNEVAQFLSKE-----------NQVIVRMR 331
Query 132 GLPWTCTEMNVVNFF 146
GLP+T T VV FF
Sbjct 332 GLPFTATAEEVVAFF 346
Score = 51.6 bits (122), Expect = 1e-06, Method: Composition-based stats.
Identities = 27/81 (33%), Positives = 47/81 (58%), Gaps = 1/81 (1%)
Query 17 IRLRGLPWDVNEDAIIRFVKPV-VEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVR 75
IRLRGLP+ + I+ F+ +IR V + + R +G+A++ + +T+ MA +
Sbjct 447 IRLRGLPYAATIEDILDFLGEFSTDIRTHGVHMVLNHQGRPSGDAFIQMKSTDRAFMAAQ 506
Query 76 MLHGRMMGARWIEVFRSSAED 96
H + M R++EVF+ SAE+
Sbjct 507 KYHKKTMKDRYVEVFQCSAEE 527
Score = 47.4 bits (111), Expect = 2e-05, Method: Composition-based stats.
Identities = 35/140 (25%), Positives = 64/140 (45%), Gaps = 12/140 (8%)
Query 17 IRLRGLPWDVNEDAIIRFVK---PVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMA 73
+R+RGLP+ + ++ F P+ +E + V + R TG+A+V E A
Sbjct 328 VRMRGLPFTATAEEVVAFFGQHCPITGGKEGILFVTYP-DGRPTGDAFVLFACEEYAQNA 386
Query 74 VRMLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGRDADVKVLNLT------- 126
+R H ++G R+IE+FRS+A + ++ R + V
Sbjct 387 LRK-HKELLGKRYIELFRSTAAEVQQVLNRFSSAPLIPLPTPPIIPVLPQQFVPPTNVRD 445
Query 127 VLKLRGLPWTCTEMNVVNFF 146
++LRGLP+ T ++++F
Sbjct 446 CIRLRGLPYAATIEDILDFL 465
Score = 32.3 bits (72), Expect = 0.68, Method: Composition-based stats.
Identities = 12/34 (35%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 126 TVLKLRGLPWTCTEMNVVNFFKSMVLRLGLTLSC 159
TV++ RGLPW ++ ++ FFK + + G C
Sbjct 225 TVVRARGLPWQSSDQDIARFFKGLNIAKGGAALC 258
> hsa:54845 ESRP1, FLJ20171, RBM35A, RMB35A; epithelial splicing
regulatory protein 1; K14947 epithelial splicing regulatory
protein 1/2
Length=677
Score = 75.9 bits (185), Expect = 7e-14, Method: Composition-based stats.
Identities = 46/135 (34%), Positives = 68/135 (50%), Gaps = 16/135 (11%)
Query 15 THIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAV 74
T +R RGLPW ++ I RF K + I + +C+ R GEA V + E RD+A+
Sbjct 225 TVVRARGLPWQSSDQDIARFFKGL-NIAKGGAALCLNAQGRRNGEALVRFVSEEHRDLAL 283
Query 75 RMLHGRMMGARWIEVFRSSAEDFER---AQQRRVAMLSSDSRDGRDADVKVLNLTVLKLR 131
+ H MG R+IEV++++ EDF + VA S N ++++R
Sbjct 284 QR-HKHHMGTRYIEVYKATGEDFLKIAGGTSNEVAQFLSKE-----------NQVIVRMR 331
Query 132 GLPWTCTEMNVVNFF 146
GLP+T T VV FF
Sbjct 332 GLPFTATAEEVVAFF 346
Score = 49.3 bits (116), Expect = 6e-06, Method: Composition-based stats.
Identities = 26/81 (32%), Positives = 46/81 (56%), Gaps = 1/81 (1%)
Query 17 IRLRGLPWDVNEDAIIRFVKP-VVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVR 75
IRLRGLP+ + I+ F+ +IR V + + R +G+A++ + + + MA +
Sbjct 447 IRLRGLPYAATIEDILDFLGEFATDIRTHGVHMVLNHQGRPSGDAFIQMKSADRAFMAAQ 506
Query 76 MLHGRMMGARWIEVFRSSAED 96
H + M R++EVF+ SAE+
Sbjct 507 KCHKKNMKDRYVEVFQCSAEE 527
Score = 47.0 bits (110), Expect = 3e-05, Method: Composition-based stats.
Identities = 35/140 (25%), Positives = 64/140 (45%), Gaps = 12/140 (8%)
Query 17 IRLRGLPWDVNEDAIIRFVK---PVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMA 73
+R+RGLP+ + ++ F P+ +E + V + R TG+A+V E A
Sbjct 328 VRMRGLPFTATAEEVVAFFGQHCPITGGKEGILFVTYP-DGRPTGDAFVLFACEEYAQNA 386
Query 74 VRMLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGRDADVKVLNLT------- 126
+R H ++G R+IE+FRS+A + ++ R + V
Sbjct 387 LRK-HKDLLGKRYIELFRSTAAEVQQVLNRFSSAPLIPLPTPPIIPVLPQQFVPPTNVRD 445
Query 127 VLKLRGLPWTCTEMNVVNFF 146
++LRGLP+ T ++++F
Sbjct 446 CIRLRGLPYAATIEDILDFL 465
Score = 32.3 bits (72), Expect = 0.68, Method: Composition-based stats.
Identities = 12/34 (35%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 126 TVLKLRGLPWTCTEMNVVNFFKSMVLRLGLTLSC 159
TV++ RGLPW ++ ++ FFK + + G C
Sbjct 225 TVVRARGLPWQSSDQDIARFFKGLNIAKGGAALC 258
> dre:572992 esrp2, FLJ20171, MGC55721, MGC77254, cb404, fa07a06,
rbm35b, sb:cb404, zgc:77254; epithelial splicing regulatory
protein 2; K14947 epithelial splicing regulatory protein
1/2
Length=736
Score = 75.5 bits (184), Expect = 8e-14, Method: Composition-based stats.
Identities = 46/135 (34%), Positives = 69/135 (51%), Gaps = 16/135 (11%)
Query 15 THIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAV 74
T IR RGLPW ++ I RF K + I + V +C+ R GEA V +E RDMA+
Sbjct 224 TVIRARGLPWQSSDQDIARFFKG-LNIAKGGVALCLNAQGRRNGEALVRFINSEHRDMAL 282
Query 75 RMLHGRMMGARWIEVFRSSAEDFER---AQQRRVAMLSSDSRDGRDADVKVLNLTVLKLR 131
H MG+R+IEV++++ E+F + VA S N ++++R
Sbjct 283 DR-HKHHMGSRYIEVYKATGEEFLKIAGGTSNEVAQFLSKE-----------NQMIIRMR 330
Query 132 GLPWTCTEMNVVNFF 146
GLP+T T +V+ F
Sbjct 331 GLPFTATPQDVLGFL 345
Score = 49.7 bits (117), Expect = 5e-06, Method: Composition-based stats.
Identities = 24/81 (29%), Positives = 47/81 (58%), Gaps = 1/81 (1%)
Query 17 IRLRGLPWDVNEDAIIRFV-KPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVR 75
IRLRGLP+ + I+ F+ + ++I+ V + + R +G+A++ + + + M +
Sbjct 452 IRLRGLPYTAAIEDILEFMGEHTIDIKPHGVHMVLNQQGRPSGDAFIQMKSADRAFMVAQ 511
Query 76 MLHGRMMGARWIEVFRSSAED 96
H +MM R++EVF+ S E+
Sbjct 512 KCHKKMMKDRYVEVFQCSTEE 532
Score = 43.1 bits (100), Expect = 4e-04, Method: Composition-based stats.
Identities = 36/160 (22%), Positives = 69/160 (43%), Gaps = 48/160 (30%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGL------NKRVTGEAYVNVHTTEMR 70
IR+RGLP+ ++ F+ P + + GL + R TG+A+V E
Sbjct 327 IRMRGLPFTATPQDVLGFLGPECPVTDG----TEGLLFVKYPDGRPTGDAFVLFACEEYA 382
Query 71 DMAVRMLHGRMMGARWIEVFRSSAEDFERAQQRRVA------------------------ 106
A++ H +++G R+IE+FRS+A + ++ R ++
Sbjct 383 QNALKK-HKQILGKRYIELFRSTAAEVQQVLNRYMSTPLISTLPPPPPPMVSVPVLATPP 441
Query 107 -MLSSDSRDGRDADVKVLNLTVLKLRGLPWTCTEMNVVNF 145
+ + ++RD ++LRGLP+T +++ F
Sbjct 442 LITTGNTRD------------CIRLRGLPYTAAIEDILEF 469
Score = 32.7 bits (73), Expect = 0.55, Method: Composition-based stats.
Identities = 12/34 (35%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 126 TVLKLRGLPWTCTEMNVVNFFKSMVLRLGLTLSC 159
TV++ RGLPW ++ ++ FFK + + G C
Sbjct 224 TVIRARGLPWQSSDQDIARFFKGLNIAKGGVALC 257
> xla:379211 esrp1, MGC53361, rbm35a; epithelial splicing regulatory
protein 1; K14947 epithelial splicing regulatory protein
1/2
Length=688
Score = 74.3 bits (181), Expect = 2e-13, Method: Composition-based stats.
Identities = 46/135 (34%), Positives = 68/135 (50%), Gaps = 16/135 (11%)
Query 15 THIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAV 74
T IR RGLPW ++ I RF K + I + +C+ R GEA V + E RD+A+
Sbjct 226 TIIRARGLPWQSSDQDIARFFKG-LNIAKGGAALCLNAQGRRNGEALVRFVSEEHRDLAL 284
Query 75 RMLHGRMMGARWIEVFRSSAEDFER---AQQRRVAMLSSDSRDGRDADVKVLNLTVLKLR 131
+ H MG R+IEV++++ EDF + VA S N ++++R
Sbjct 285 QR-HKHHMGNRYIEVYKATGEDFLKIAGGTSNEVAQFLSKE-----------NQVIVRMR 332
Query 132 GLPWTCTEMNVVNFF 146
GLP+T T V+ FF
Sbjct 333 GLPFTATAEEVLAFF 347
Score = 48.9 bits (115), Expect = 7e-06, Method: Composition-based stats.
Identities = 30/98 (30%), Positives = 52/98 (53%), Gaps = 6/98 (6%)
Query 2 LPTPASNGIPGVGTH--IRLRGLPWDVNEDAIIRFVKPV-VEIRESDVCVCVGLNKRVTG 58
LP P IP V IRLRGLP+ + I+ F+ +IR V + + R +G
Sbjct 434 LPQPF---IPPVNVRDCIRLRGLPYAATIEDILEFLGEFSADIRTHGVHMVLNHQGRPSG 490
Query 59 EAYVNVHTTEMRDMAVRMLHGRMMGARWIEVFRSSAED 96
++++ + + + +A + H + M R++EVF+ SAE+
Sbjct 491 DSFIQMKSADRAYLAAQKCHKKTMKDRYVEVFQCSAEE 528
Score = 46.6 bits (109), Expect = 3e-05, Method: Composition-based stats.
Identities = 37/140 (26%), Positives = 64/140 (45%), Gaps = 12/140 (8%)
Query 17 IRLRGLPWDVNEDAIIRFVK---PVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMA 73
+R+RGLP+ + ++ F PV +E + V N R TG+A+V E A
Sbjct 329 VRMRGLPFTATAEEVLAFFGQQCPVTGGKEGILFVTYPDN-RPTGDAFVLFACEEYAQNA 387
Query 74 VRMLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGRDADV-------KVLNLT 126
++ H ++G R+IE+FRS+A + ++ R + V V
Sbjct 388 LKK-HKELLGKRYIELFRSTAAEVQQVLNRYSSAPLIPLPTPPIIPVLPQPFIPPVNVRD 446
Query 127 VLKLRGLPWTCTEMNVVNFF 146
++LRGLP+ T +++ F
Sbjct 447 CIRLRGLPYAATIEDILEFL 466
Score = 32.3 bits (72), Expect = 0.79, Method: Composition-based stats.
Identities = 11/34 (32%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 126 TVLKLRGLPWTCTEMNVVNFFKSMVLRLGLTLSC 159
T+++ RGLPW ++ ++ FFK + + G C
Sbjct 226 TIIRARGLPWQSSDQDIARFFKGLNIAKGGAALC 259
> dre:560190 esrp1, rbm35a, wu:fi28a07, zgc:154050; epithelial
splicing regulatory protein 1; K14947 epithelial splicing regulatory
protein 1/2
Length=714
Score = 73.6 bits (179), Expect = 3e-13, Method: Composition-based stats.
Identities = 45/133 (33%), Positives = 70/133 (52%), Gaps = 12/133 (9%)
Query 15 THIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAV 74
T IR RGLPW ++ I RF + + I + +C+ R GEA V + E RD+A+
Sbjct 225 TVIRARGLPWQSSDQDIARFFRGL-NIAKGGAALCLNAQGRRNGEALVRFESEEHRDLAL 283
Query 75 RMLHGRMMGARWIEVFRSSAEDFER-AQQRRVAMLSSDSRDGRDADVKVLNLTVLKLRGL 133
+ H MG R+IEV++++ EDF + A + S SR+ N ++++RGL
Sbjct 284 QR-HKHHMGGRYIEVYKATGEDFLKIAGGTSNEVASFLSRE---------NQIIVRMRGL 333
Query 134 PWTCTEMNVVNFF 146
P+ T V+ FF
Sbjct 334 PFNATAEQVLQFF 346
Score = 59.7 bits (143), Expect = 5e-09, Method: Composition-based stats.
Identities = 31/97 (31%), Positives = 53/97 (54%), Gaps = 1/97 (1%)
Query 1 PLPTPASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPV-VEIRESDVCVCVGLNKRVTGE 59
P P + +PGV +RLRGLP+D + I+ F+ +I+ V + + R +GE
Sbjct 436 PTFVPQTAAVPGVRDCVRLRGLPYDASIQDILVFLGEYGADIKTHGVHMVLNHQGRPSGE 495
Query 60 AYVNVHTTEMRDMAVRMLHGRMMGARWIEVFRSSAED 96
A++ + + E +A + H R M R++EVF SA++
Sbjct 496 AFIQMRSAERAFLAAQRCHKRSMKERYVEVFACSAQE 532
Score = 45.8 bits (107), Expect = 7e-05, Method: Composition-based stats.
Identities = 25/85 (29%), Positives = 50/85 (58%), Gaps = 3/85 (3%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIRE-SDVCVCVGL-NKRVTGEAYVNVHTTEMRDMAV 74
+R+RGLP++ + +++F P + + S+ + V + R TG+A+V E A+
Sbjct 328 VRMRGLPFNATAEQVLQFFSPACPVTDGSEGILFVRFPDGRPTGDAFVLFSCEEHAQNAL 387
Query 75 RMLHGRMMGARWIEVFRSSAEDFER 99
+ H M+G R+IE+F+S+A + ++
Sbjct 388 KK-HKDMLGKRYIELFKSTAAEVQQ 411
Score = 32.0 bits (71), Expect = 1.0, Method: Composition-based stats.
Identities = 12/39 (30%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 121 KVLNLTVLKLRGLPWTCTEMNVVNFFKSMVLRLGLTLSC 159
+V + TV++ RGLPW ++ ++ FF+ + + G C
Sbjct 220 RVDDNTVIRARGLPWQSSDQDIARFFRGLNIAKGGAALC 258
> mmu:77411 Esrp2, 9530027K23Rik, Rbm35b; epithelial splicing
regulatory protein 2; K14947 epithelial splicing regulatory
protein 1/2
Length=717
Score = 67.4 bits (163), Expect = 2e-11, Method: Composition-based stats.
Identities = 41/133 (30%), Positives = 70/133 (52%), Gaps = 12/133 (9%)
Query 15 THIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAV 74
T +R RGLPW ++ + RF K + I V +C+ R GEA + +E RD+A+
Sbjct 247 TVVRARGLPWQSSDQDVARFFKG-LNIARGGVALCLNAQGRRNGEALIRFVDSEQRDLAL 305
Query 75 RMLHGRMMGARWIEVFRSSAEDFER-AQQRRVAMLSSDSRDGRDADVKVLNLTVLKLRGL 133
+ H MG R+IEV++++ E+F + A + + SR+ + +L+LRGL
Sbjct 306 QR-HKHHMGVRYIEVYKATGEEFVKIAGGTSLEVARFLSREDQ---------VILRLRGL 355
Query 134 PWTCTEMNVVNFF 146
P++ +V+ F
Sbjct 356 PFSAGPTDVLGFL 368
Score = 49.7 bits (117), Expect = 4e-06, Method: Composition-based stats.
Identities = 31/100 (31%), Positives = 51/100 (51%), Gaps = 6/100 (6%)
Query 17 IRLRGLPWDVNEDAIIRFV-KPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVR 75
+RLRGLP+ + I+ F+ + +IR V + + R +G+A++ + + E A +
Sbjct 467 VRLRGLPYTATIEDILSFLGEAAADIRPHGVHMVLNQQGRPSGDAFIQMMSVERALAAAQ 526
Query 76 MLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDG 115
H +MM R++EV S E+ RV M S SR G
Sbjct 527 RCHKKMMKERYVEVVPCSTEEMS-----RVLMGGSLSRSG 561
Score = 49.3 bits (116), Expect = 6e-06, Method: Composition-based stats.
Identities = 38/146 (26%), Positives = 65/146 (44%), Gaps = 26/146 (17%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIR--ESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAV 74
+RLRGLP+ ++ F+ P + + + R TG+A+ E+ A+
Sbjct 350 LRLRGLPFSAGPTDVLGFLGPECPVTGGADGLLFVRHPDGRPTGDAFALFACEELAQAAL 409
Query 75 RMLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSR--------------DGRDADV 120
R H M+G R+IE+FRS+A + ++ R A + GRD
Sbjct 410 RR-HKGMLGKRYIELFRSTAAEVQQVLNRYAASPLLPTLTAPLLPIPFPLAGGTGRDC-- 466
Query 121 KVLNLTVLKLRGLPWTCTEMNVVNFF 146
++LRGLP+T T ++++F
Sbjct 467 -------VRLRGLPYTATIEDILSFL 485
Score = 35.0 bits (79), Expect = 0.11, Method: Composition-based stats.
Identities = 19/41 (46%), Positives = 27/41 (65%), Gaps = 2/41 (4%)
Query 118 ADVKVLNLTVLKLRGLPWTCTEMNVVNFFKSM-VLRLGLTL 157
ADV V N TV++ RGLPW ++ +V FFK + + R G+ L
Sbjct 240 ADV-VDNETVVRARGLPWQSSDQDVARFFKGLNIARGGVAL 279
> hsa:80004 ESRP2, FLJ21918, FLJ22248, RBM35B; epithelial splicing
regulatory protein 2; K14947 epithelial splicing regulatory
protein 1/2
Length=717
Score = 66.2 bits (160), Expect = 4e-11, Method: Composition-based stats.
Identities = 40/133 (30%), Positives = 70/133 (52%), Gaps = 12/133 (9%)
Query 15 THIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAV 74
T +R RGLPW ++ + RF K + + V +C+ R GEA + +E RD+A+
Sbjct 247 TVVRARGLPWQSSDQDVARFFKG-LNVARGGVALCLNAQGRRNGEALIRFVDSEQRDLAL 305
Query 75 RMLHGRMMGARWIEVFRSSAEDFER-AQQRRVAMLSSDSRDGRDADVKVLNLTVLKLRGL 133
+ H MG R+IEV++++ E+F + A + + SR+ + +L+LRGL
Sbjct 306 QR-HKHHMGVRYIEVYKATGEEFVKIAGGTSLEVARFLSREDQ---------VILRLRGL 355
Query 134 PWTCTEMNVVNFF 146
P++ +V+ F
Sbjct 356 PFSAGPTDVLGFL 368
Score = 48.1 bits (113), Expect = 1e-05, Method: Composition-based stats.
Identities = 29/100 (29%), Positives = 52/100 (52%), Gaps = 5/100 (5%)
Query 2 LPTPASNGIPGVGTH-IRLRGLPWDVNEDAIIRFV-KPVVEIRESDVCVCVGLNKRVTGE 59
+P P + PG G +RLRGLP+ + I+ F+ + +IR V + + R +G+
Sbjct 454 IPFPLA---PGTGRDCVRLRGLPYTATIEDILSFLGEAAADIRPHGVHMVLNQQGRPSGD 510
Query 60 AYVNVHTTEMRDMAVRMLHGRMMGARWIEVFRSSAEDFER 99
A++ + + E A + H ++M R++EV S E+ R
Sbjct 511 AFIQMTSAERALAAAQRCHKKVMKERYVEVVPCSTEEMSR 550
Score = 47.4 bits (111), Expect = 2e-05, Method: Composition-based stats.
Identities = 37/146 (25%), Positives = 65/146 (44%), Gaps = 26/146 (17%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIR--ESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAV 74
+RLRGLP+ ++ F+ P + + + R TG+A+ E+ A+
Sbjct 350 LRLRGLPFSAGPTDVLGFLGPECPVTGGTEGLLFVRHPDGRPTGDAFALFACEELAQAAL 409
Query 75 RMLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSR--------------DGRDADV 120
R H M+G R+IE+FRS+A + ++ R + + GRD
Sbjct 410 RR-HKGMLGKRYIELFRSTAAEVQQVLNRYASGPLLPTLTAPLLPIPFPLAPGTGRDC-- 466
Query 121 KVLNLTVLKLRGLPWTCTEMNVVNFF 146
++LRGLP+T T ++++F
Sbjct 467 -------VRLRGLPYTATIEDILSFL 485
Score = 33.5 bits (75), Expect = 0.36, Method: Composition-based stats.
Identities = 19/41 (46%), Positives = 27/41 (65%), Gaps = 2/41 (4%)
Query 118 ADVKVLNLTVLKLRGLPWTCTEMNVVNFFKSM-VLRLGLTL 157
ADV V + TV++ RGLPW ++ +V FFK + V R G+ L
Sbjct 240 ADV-VDSETVVRARGLPWQSSDQDVARFFKGLNVARGGVAL 279
> cel:Y73B6BL.33 hypothetical protein; K12898 heterogeneous nuclear
ribonucleoprotein F/H
Length=610
Score = 66.2 bits (160), Expect = 5e-11, Method: Composition-based stats.
Identities = 48/148 (32%), Positives = 72/148 (48%), Gaps = 18/148 (12%)
Query 2 LPTPASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAY 61
+PT S+G P +IRLRGLP++ E I F + R VC R GEAY
Sbjct 51 MPTKISSGEPPRSQYIRLRGLPFNATEKDIHEFFAGLTIERVKFVCT----TGRPNGEAY 106
Query 62 VNVHTTEMRDMAVRMLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGRDADVK 121
V TE A+ + + R+IEVF A++ E + R D++ +
Sbjct 107 VEFKNTEEAGKAMEN-DRKEISNRYIEVFTVEADEAE-----------FEFRPDPDSNGE 154
Query 122 VLNLTVLKLRGLPWTCTEMNVVNFFKSM 149
V + V++LRG+PW+C E +V FF+ +
Sbjct 155 VNH--VIRLRGVPWSCKEDDVRKFFEGL 180
Score = 58.5 bits (140), Expect = 9e-09, Method: Composition-based stats.
Identities = 37/101 (36%), Positives = 57/101 (56%), Gaps = 6/101 (5%)
Query 3 PTPASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCV--CVGLNKRVTGEA 60
P P SNG V IRLRG+PW ED + +F + + E +++ + G R +GEA
Sbjct 147 PDPDSNG--EVNHVIRLRGVPWSCKEDDVRKFFEGL-EPPPAEIVIGGTGGPRSRPSGEA 203
Query 61 YVNVHTTEMRDMAVRMLHGRMMGARWIEVFRSSAEDFERAQ 101
+V T + + A+ + R MG+R++EVF SS +F RA+
Sbjct 204 FVRFTTQDAAEKAMDY-NNRHMGSRYVEVFMSSMVEFNRAK 243
Score = 33.5 bits (75), Expect = 0.32, Method: Composition-based stats.
Identities = 21/74 (28%), Positives = 37/74 (50%), Gaps = 4/74 (5%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVRM 76
+ +RGLP+D ++ AI F P +R V + + R +G+A +A+
Sbjct 327 VYMRGLPYDADDHAIAAFFSP---LRCHSVKIGINETGRPSGDAIAEFDNYNDLQVALSR 383
Query 77 LHGRMMGARWIEVF 90
+ R MG R++E+F
Sbjct 384 NNQR-MGRRYVELF 396
> cel:ZK1067.6 sym-2; SYnthetic lethal with Mec family member
(sym-2); K14947 epithelial splicing regulatory protein 1/2
Length=618
Score = 63.2 bits (152), Expect = 4e-10, Method: Composition-based stats.
Identities = 39/143 (27%), Positives = 70/143 (48%), Gaps = 11/143 (7%)
Query 9 GIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTE 68
G G R RGLPW ++ + +F + +I + +C+ R GE V + E
Sbjct 175 GADGDNVVCRARGLPWQASDHHVAQFFAGL-DIVPGGIALCLSSEGRRNGEVLVQFSSQE 233
Query 69 MRDMAVRMLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGRDADVKVLNLTVL 128
RD+A++ H + +R+IEV+++ ++F VA SS + N ++
Sbjct 234 SRDLALKR-HRNFLLSRYIEVYKAGLDEF-----MHVATGSSTEA----MEFVSANAIIV 283
Query 129 KLRGLPWTCTEMNVVNFFKSMVL 151
++RGLP+ CT+ + FF+ + L
Sbjct 284 RMRGLPYDCTDAQIRTFFEPLKL 306
Score = 57.0 bits (136), Expect = 3e-08, Method: Composition-based stats.
Identities = 39/138 (28%), Positives = 73/138 (52%), Gaps = 8/138 (5%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIRESD-VCVCVGLNKRVTGEAYVNVHTTEMRDMAVR 75
+R+RGLP+D + I F +P ++ +D + + R TG+A+V T E +
Sbjct 283 VRMRGLPYDCTDAQIRTFFEP---LKLTDKILFITRTDGRPTGDAFVQFETEEDAQQGL- 338
Query 76 MLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGRDADVKVLNLTVLKLRGLPW 135
+ H +++G R+IE+F+S+A + ++ +R + SS + + ++LRGLP+
Sbjct 339 LKHRQVIGQRYIELFKSTAAEVQQVVKRCNLINSSPAVANAVEAPEEKKKDCVRLRGLPY 398
Query 136 TCTEMNVVNF---FKSMV 150
T ++V F F +MV
Sbjct 399 EATVQHIVTFLGDFATMV 416
Score = 35.8 bits (81), Expect = 0.068, Method: Composition-based stats.
Identities = 22/86 (25%), Positives = 40/86 (46%), Gaps = 5/86 (5%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVE-IRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVR 75
+RLRGLP++ I+ F+ ++ V + +GEA++ + +
Sbjct 391 VRLRGLPYEATVQHIVTFLGDFATMVKFQGVHMVYNNQGHPSGEAFIQMINEQAASACAA 450
Query 76 MLHGRMMGA----RWIEVFRSSAEDF 97
+H M R+IEVF++SAE+
Sbjct 451 GVHNNFMSVGKKKRYIEVFQASAEEL 476
> xla:100158276 grsf1; G-rich RNA sequence binding factor 1
Length=348
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 52/204 (25%), Positives = 88/204 (43%), Gaps = 50/204 (24%)
Query 4 TPASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVN 63
+P S+GI +RLRGLP+ +E II F + +I + + + R +GEA+V
Sbjct 113 SPPSDGI------VRLRGLPYSCSEQDIIHFFSGL-DIADEGITFVLDQRGRKSGEAFVQ 165
Query 64 VHTTEMRDMAVRMLHGRMMGARWIEVFRSSAEDFERA----QQRRVAMLSSDSRDGRDAD 119
+ E D A+ + H + +G+R+IE+F S D + A ++R+ + +D D D
Sbjct 166 FLSQEHADQAL-LKHKQEIGSRYIEIFPSRRNDVQTARFPFRRRKGVTFAPTIKDLYDPD 224
Query 120 -------------------------------VKVLNLTVL------KLRGLPWTCTEMNV 142
V V + TV+ +RGLP+ + ++
Sbjct 225 NCINNTSKDLLSDVPENGHINDYVKEMSAKSVDVHDFTVMSPVHDIHIRGLPFHASGQDI 284
Query 143 VNFFKS-MVLRLGLTLSCWASRWT 165
NFF M L++ + S A T
Sbjct 285 ANFFHPIMPLKISIEYSADAGGAT 308
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 38/149 (25%), Positives = 70/149 (46%), Gaps = 10/149 (6%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNK--RVTGEAYVNVHTTEMRDMAV 74
+R+RGLPW D ++ F +R V N+ + G+A + + E AV
Sbjct 4 VRVRGLPWSCTADDVLNFFDDS-NVRNGTDGVHFIFNRDGKPRGDAVIEFESAEDVQKAV 62
Query 75 RMLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGRDADVKVLNLT-----VLK 129
H + MG R++EVF + ++ E R + LS + + +++
Sbjct 63 EQ-HKKYMGQRYVEVFEMNQKEAESLLNRMHSALSPTRPSSMSLSPQSSMASPPSDGIVR 121
Query 130 LRGLPWTCTEMNVVNFFKSM-VLRLGLTL 157
LRGLP++C+E ++++FF + + G+T
Sbjct 122 LRGLPYSCSEQDIIHFFSGLDIADEGITF 150
Score = 37.0 bits (84), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 125 LTVLKLRGLPWTCTEMNVVNFFKSMVLRLG 154
+ ++++RGLPW+CT +V+NFF +R G
Sbjct 1 MFIVRVRGLPWSCTADDVLNFFDDSNVRNG 30
> dre:321556 hnrnph1l, fb19a06, wu:fb19a06, zgc:55399, zgc:85960;
heterogeneous nuclear ribonucleoprotein H1, like; K12898
heterogeneous nuclear ribonucleoprotein F/H
Length=407
Score = 59.3 bits (142), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 41/171 (23%), Positives = 74/171 (43%), Gaps = 40/171 (23%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVRM 76
+RLRGLP+ +++ I++F +EI + + + V R TGEA+V + ++ + A++
Sbjct 109 VRLRGLPFGCSKEEIVQFFS-GLEIVPNGITLPVDFQGRSTGEAFVQFASQDIAEKALKK 167
Query 77 LHGRMMGARWIEVFRSS--------------------------------------AEDFE 98
H +G R+IE+F+SS F+
Sbjct 168 -HKERIGHRYIEIFKSSRAEVRTHYEPPRKGMGMQRPGPYDRPSGGGRGYNGMSRGGSFD 226
Query 99 RAQQRRVAMLSSDSRDGRDADVKVLNLTVLKLRGLPWTCTEMNVVNFFKSM 149
R ++ SD R G + + + +RGLP+ TE ++ NFF +
Sbjct 227 RMRRGGYGGGVSDGRYGDGGNFQSTTGHCVHMRGLPYRATEPDIYNFFSPL 277
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 37/143 (25%), Positives = 69/143 (48%), Gaps = 14/143 (9%)
Query 17 IRLRGLPWDVNEDAIIRFVKPV-VEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVR 75
+R+RGLPW + + + RF + S + R +GEA+V + + + +AV+
Sbjct 9 VRVRGLPWSCSAEEVSRFFSGCKISSNGSAIHFTYTREGRPSGEAFVELESEDDLKIAVK 68
Query 76 MLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGRDADVKVLNLTVLKLRGLPW 135
MG R++EVF+S+ + + + DG +++LRGLP+
Sbjct 69 KDR-ESMGHRYVEVFKSNNVEMDWVLKHTGPNCPETEGDG-----------LVRLRGLPF 116
Query 136 TCTEMNVVNFFKSM-VLRLGLTL 157
C++ +V FF + ++ G+TL
Sbjct 117 GCSKEEIVQFFSGLEIVPNGITL 139
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 28/82 (34%), Positives = 42/82 (51%), Gaps = 6/82 (7%)
Query 13 VGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDM 72
G + +RGLP+ E I F P+ +R V + +G + RVTGEA V T E D
Sbjct 252 TGHCVHMRGLPYRATEPDIYNFFSPLNPVR---VHIEIGPDGRVTGEADVEFATHE--DA 306
Query 73 AVRMLHGRM-MGARWIEVFRSS 93
M + + M R++E+F +S
Sbjct 307 VAAMSNDKANMQHRYVELFLNS 328
Score = 30.0 bits (66), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 10/20 (50%), Positives = 15/20 (75%), Gaps = 0/20 (0%)
Query 127 VLKLRGLPWTCTEMNVVNFF 146
V+++RGLPW+C+ V FF
Sbjct 8 VVRVRGLPWSCSAEEVSRFF 27
> hsa:3188 HNRNPH2, FTP3, HNRPH', HNRPH2, hnRNPH'; heterogeneous
nuclear ribonucleoprotein H2 (H'); K12898 heterogeneous nuclear
ribonucleoprotein F/H
Length=449
Score = 58.5 bits (140), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 37/143 (25%), Positives = 72/143 (50%), Gaps = 14/143 (9%)
Query 17 IRLRGLPWDVNEDAIIRFVKPV-VEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVR 75
+++RGLPW + D ++RF ++ S + R +GEA+V + + E +A++
Sbjct 13 VKVRGLPWSCSADEVMRFFSDCKIQNGTSGIRFIYTREGRPSGEAFVELESEEEVKLALK 72
Query 76 MLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGRDADVKVLNLTVLKLRGLPW 135
MG R++EVF+S++ + + + + DG ++LRGLP+
Sbjct 73 KDR-ETMGHRYVEVFKSNSVEMDWVLKHTGPNSPDTANDG-----------FVRLRGLPF 120
Query 136 TCTEMNVVNFFKSM-VLRLGLTL 157
C++ +V FF + ++ G+TL
Sbjct 121 GCSKEEIVQFFSGLEIVPNGMTL 143
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 30/91 (32%), Positives = 53/91 (58%), Gaps = 8/91 (8%)
Query 3 PTPASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYV 62
P A++G +RLRGLP+ +++ I++F + EI + + + V R TGEA+V
Sbjct 105 PDTANDGF------VRLRGLPFGCSKEEIVQFFSGL-EIVPNGMTLPVDFQGRSTGEAFV 157
Query 63 NVHTTEMRDMAVRMLHGRMMGARWIEVFRSS 93
+ E+ + A++ H +G R+IE+F+SS
Sbjct 158 QFASQEIAEKALKK-HKERIGHRYIEIFKSS 187
Score = 42.4 bits (98), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 28/82 (34%), Positives = 42/82 (51%), Gaps = 6/82 (7%)
Query 13 VGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDM 72
G + +RGLP+ E+ I F P+ +R V + +G + RVTGEA V T E D
Sbjct 287 TGHCVHMRGLPYRATENDIYNFFSPLNPMR---VHIEIGPDGRVTGEADVEFATHE--DA 341
Query 73 AVRMLHGRM-MGARWIEVFRSS 93
M + M R++E+F +S
Sbjct 342 VAAMAKDKANMQHRYVELFLNS 363
Score = 35.0 bits (79), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 28/50 (56%), Gaps = 10/50 (20%)
Query 107 MLSSDSRDGRDADVKVLNLTVLKLRGLPWTCTEMNVVNFFKSMVLRLGLT 156
MLS++ R+G V+K+RGLPW+C+ V+ FF ++ G +
Sbjct 2 MLSTEGREG----------FVVKVRGLPWSCSADEVMRFFSDCKIQNGTS 41
> dre:402984 hnrnph1, MGC77712, zgc:77712; heterogeneous nuclear
ribonucleoprotein H1; K12898 heterogeneous nuclear ribonucleoprotein
F/H
Length=403
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 40/148 (27%), Positives = 71/148 (47%), Gaps = 14/148 (9%)
Query 12 GVGTHIRLRGLPWDVNEDAIIRFVKPV-VEIRESDVCVCVGLNKRVTGEAYVNVHTTEMR 70
G G +R+RGLPW + D + RF + + + R +GEA+V + E
Sbjct 4 GEGFVVRVRGLPWSCSVDEVQRFFSECKIASNGTSIHFTYTREGRPSGEAFVEFESEEDL 63
Query 71 DMAVRMLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGRDADVKVLNLTVLKL 130
+AV+ MG R++EVF+S++ + + + DG +++L
Sbjct 64 KIAVKKDR-ETMGHRYVEVFKSNSVEMDWVLKHTGPNCPETGGDG-----------LVRL 111
Query 131 RGLPWTCTEMNVVNFFKSM-VLRLGLTL 157
RGLP+ C++ +V FF + ++ G+TL
Sbjct 112 RGLPFGCSKEEIVQFFAGLEIVPNGITL 139
Score = 56.2 bits (134), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 41/168 (24%), Positives = 73/168 (43%), Gaps = 37/168 (22%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVRM 76
+RLRGLP+ +++ I++F + EI + + + V R TGEA+V + ++ + A++
Sbjct 109 VRLRGLPFGCSKEEIVQFFAGL-EIVPNGITLPVDFQGRSTGEAFVQFASQDIAEKALKK 167
Query 77 LHGRMMGARWIEVFRSSAEDF--ERAQQRRVAMLSSDSRDGRDADVKVLNL--------- 125
H +G R+IE+F+SS + QR+V + S R + N
Sbjct 168 -HKERIGHRYIEIFKSSRAEVRTHYEPQRKVMGMQRPSPYDRPGGGRGYNSMGRGVSFER 226
Query 126 ------------------------TVLKLRGLPWTCTEMNVVNFFKSM 149
+ +RGLP+ TE ++ NFF +
Sbjct 227 MRRGGYGGDGRYGDSGSSFQSTTGHCVHMRGLPYRATETDIYNFFSPL 274
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 28/82 (34%), Positives = 41/82 (50%), Gaps = 6/82 (7%)
Query 13 VGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDM 72
G + +RGLP+ E I F P+ +R V + +G + RVTGEA V T E D
Sbjct 249 TGHCVHMRGLPYRATETDIYNFFSPLNPVR---VHLEIGPDGRVTGEADVEFATHE--DA 303
Query 73 AVRMLHGRM-MGARWIEVFRSS 93
M + M R++E+F +S
Sbjct 304 VAAMSKDKANMQHRYVELFLNS 325
> hsa:3185 HNRNPF, HNRPF, MGC110997, OK/SW-cl.23, mcs94-1; heterogeneous
nuclear ribonucleoprotein F; K12898 heterogeneous
nuclear ribonucleoprotein F/H
Length=415
Score = 56.2 bits (134), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 40/148 (27%), Positives = 71/148 (47%), Gaps = 14/148 (9%)
Query 12 GVGTHIRLRGLPWDVNEDAIIRFVKP-VVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMR 70
G G ++LRGLPW + + + F+ + + V R +GEA+V + + +
Sbjct 8 GEGFVVKLRGLPWSCSVEDVQNFLSDCTIHDGAAGVHFIYTREGRQSGEAFVELGSEDDV 67
Query 71 DMAVRMLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGRDADVKVLNLTVLKL 130
MA++ MG R+IEVF+S + + + + + DG ++L
Sbjct 68 KMALKKDR-ESMGHRYIEVFKSHRTEMDWVLKHSGPNSADSANDG-----------FVRL 115
Query 131 RGLPWTCTEMNVVNFFKSM-VLRLGLTL 157
RGLP+ CT+ +V FF + ++ G+TL
Sbjct 116 RGLPFGCTKEEIVQFFSGLEIVPNGITL 143
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 49/80 (61%), Gaps = 2/80 (2%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVRM 76
+RLRGLP+ ++ I++F + EI + + + V ++TGEA+V + E+ + A+
Sbjct 113 VRLRGLPFGCTKEEIVQFFSGL-EIVPNGITLPVDPEGKITGEAFVQFASQELAEKALGK 171
Query 77 LHGRMMGARWIEVFRSSAED 96
H +G R+IEVF+SS E+
Sbjct 172 -HKERIGHRYIEVFKSSQEE 190
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 41/81 (50%), Gaps = 4/81 (4%)
Query 13 VGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDM 72
G + +RGLP+ E+ I F P+ +R V + +G + RVTGEA V T E
Sbjct 287 TGHCVHMRGLPYKATENDIYNFFSPLNPVR---VHIEIGPDGRVTGEADVEFATHEEAVA 343
Query 73 AVRMLHGRMMGARWIEVFRSS 93
A+ M R+IE+F +S
Sbjct 344 AMSKDRANMQ-HRYIELFLNS 363
> mmu:56258 Hnrnph2, DXHXS1271E, Ftp-3, Ftp3, H', HNRNP, Hnrph2;
heterogeneous nuclear ribonucleoprotein H2; K12898 heterogeneous
nuclear ribonucleoprotein F/H
Length=449
Score = 55.8 bits (133), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 36/143 (25%), Positives = 72/143 (50%), Gaps = 14/143 (9%)
Query 17 IRLRGLPWDVNEDAIIRFVKPV-VEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVR 75
+++RGLPW + + ++RF ++ S V R +GEA+V + + + +A++
Sbjct 13 VKVRGLPWSCSAEEVMRFFSDCKIQNGTSGVRFIYTREGRPSGEAFVELESEDEVKLALK 72
Query 76 MLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGRDADVKVLNLTVLKLRGLPW 135
MG R++EVF+S++ + + + + DG ++LRGLP+
Sbjct 73 KDR-ETMGHRYVEVFKSNSVEMDWVLKHTGPNSPDTANDG-----------FVRLRGLPF 120
Query 136 TCTEMNVVNFFKSM-VLRLGLTL 157
C++ +V FF + ++ G+TL
Sbjct 121 GCSKEEIVQFFSGLEIVPNGMTL 143
Score = 53.5 bits (127), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 30/91 (32%), Positives = 53/91 (58%), Gaps = 8/91 (8%)
Query 3 PTPASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYV 62
P A++G +RLRGLP+ +++ I++F + EI + + + V R TGEA+V
Sbjct 105 PDTANDGF------VRLRGLPFGCSKEEIVQFFSGL-EIVPNGMTLPVDFQGRSTGEAFV 157
Query 63 NVHTTEMRDMAVRMLHGRMMGARWIEVFRSS 93
+ E+ + A++ H +G R+IE+F+SS
Sbjct 158 QFASQEIAEKALKK-HKERIGHRYIEIFKSS 187
Score = 42.4 bits (98), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 28/82 (34%), Positives = 42/82 (51%), Gaps = 6/82 (7%)
Query 13 VGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDM 72
G + +RGLP+ E+ I F P+ +R V + +G + RVTGEA V T E D
Sbjct 287 TGHCVHMRGLPYRATENDIYNFFSPLNPMR---VHIEIGPDGRVTGEADVEFATHE--DA 341
Query 73 AVRMLHGRM-MGARWIEVFRSS 93
M + M R++E+F +S
Sbjct 342 VAAMAKDKANMQHRYVELFLNS 363
Score = 35.4 bits (80), Expect = 0.092, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 28/50 (56%), Gaps = 10/50 (20%)
Query 107 MLSSDSRDGRDADVKVLNLTVLKLRGLPWTCTEMNVVNFFKSMVLRLGLT 156
MLS++ R+G V+K+RGLPW+C+ V+ FF ++ G +
Sbjct 2 MLSTEGREG----------FVVKVRGLPWSCSAEEVMRFFSDCKIQNGTS 41
> ath:AT5G66010 RNA binding / nucleic acid binding / nucleotide
binding
Length=255
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 44/142 (30%), Positives = 72/142 (50%), Gaps = 18/142 (12%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAV-R 75
+RLRGLP++ + I F + + DV + V N + +GEA+V ++A+ R
Sbjct 51 VRLRGLPFNCADIDIFEFFAGLNIV---DVLL-VSKNGKFSGEAFVVFAGPMQVEIALQR 106
Query 76 MLHGRMMGARWIEVFRSSAEDFERA----------QQRRVAMLSSDSRDGRDADVKVLNL 125
H MG R++EVFR S +D+ A + R + SR R ++ + L
Sbjct 107 DRHN--MGRRYVEVFRCSKQDYYNAVAAEEGAYEYEVRASPPPTGPSRAKRFSEKEKLEY 164
Query 126 T-VLKLRGLPWTCTEMNVVNFF 146
T VLK+RGLP++ + ++ FF
Sbjct 165 TEVLKMRGLPYSVNKPQIIEFF 186
Score = 42.4 bits (98), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 28/77 (36%), Positives = 43/77 (55%), Gaps = 8/77 (10%)
Query 17 IRLRGLPWDVNEDAIIRFVK--PVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAV 74
+++RGLP+ VN+ II F V++ R VC G + TGEA+V T E A
Sbjct 168 LKMRGLPYSVNKPQIIEFFSGYKVIQGRVQVVCRPDG---KATGEAFVEFETGEEARRA- 223
Query 75 RMLHGRM-MGARWIEVF 90
M +M +G+R++E+F
Sbjct 224 -MAKDKMSIGSRYVELF 239
> dre:563249 grsf1, MGC153305, wu:fb62c04, zgc:153305; G-rich
RNA sequence binding factor 1
Length=301
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 41/141 (29%), Positives = 71/141 (50%), Gaps = 9/141 (6%)
Query 15 THIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAV 74
T +RLRGLP+ E IIRF +++ E V + + + +G+A+V T M + A+
Sbjct 134 TVVRLRGLPYSCTEGDIIRFFS-GLDVVEDGVTIILNRRGKSSGDAFVEFATKAMAEKAL 192
Query 75 RMLHGRMMGARWIEVF---RSSAEDFERA-QQRRVAMLSSDSRDGRDADVKVLNLTVLKL 130
+ ++G R+IE+F +S+ R+ Q RV ++ R+ V V+ +
Sbjct 193 KKDR-EILGNRYIEIFPAMKSAIPSQNRSWQNDRVFTPRAEDPPLRNTAV---TKNVIHM 248
Query 131 RGLPWTCTEMNVVNFFKSMVL 151
RGLP+ ++V FF + L
Sbjct 249 RGLPFDAKAEDIVKFFAPVRL 269
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 47/163 (28%), Positives = 75/163 (46%), Gaps = 22/163 (13%)
Query 1 PLP--TPASNGIPGVGTHI-RLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNK--R 55
PLP TP S I + +GLPW + I+ F IR V + NK +
Sbjct 20 PLPEYTPGSEENQAKELFIVQAKGLPWSCTAEDIMSFFSEC-RIRGGVNGVHILYNKYGK 78
Query 56 VTGEAYVNVHTTEMRDMAVRMLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDG 115
+G+A++ + E A+ H R IEV + +D E A+L +
Sbjct 79 PSGQAFIELEHEEDVGKALDQ-HRHYPRDRLIEVREVTNKDAE-------AILKASKE-- 128
Query 116 RDADVKVLNLTVLKLRGLPWTCTEMNVVNFFKSM-VLRLGLTL 157
+V TV++LRGLP++CTE +++ FF + V+ G+T+
Sbjct 129 -----RVETDTVVRLRGLPYSCTEGDIIRFFSGLDVVEDGVTI 166
Score = 35.0 bits (79), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 26/52 (50%), Gaps = 3/52 (5%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTE 68
I +RGLP+D + I++F PV R V V G + TGEA T E
Sbjct 246 IHMRGLPFDAKAEDIVKFFAPV---RLMKVVVEFGPEGKPTGEAEAYFKTHE 294
> mmu:98758 Hnrnpf, 4833420I20Rik, AA407306, Hnrpf, MGC101981,
MGC36971; heterogeneous nuclear ribonucleoprotein F; K12898
heterogeneous nuclear ribonucleoprotein F/H
Length=415
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 39/148 (26%), Positives = 71/148 (47%), Gaps = 14/148 (9%)
Query 12 GVGTHIRLRGLPWDVNEDAIIRFVKP-VVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMR 70
G G ++LRGLPW + + + F+ + + V R +GEA+V + + +
Sbjct 8 GEGYVVKLRGLPWSCSIEDVQNFLSDCTIHDGVAGVHFIYTREGRQSGEAFVELESEDDV 67
Query 71 DMAVRMLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGRDADVKVLNLTVLKL 130
+A++ MG R+IEVF+S + + + + + DG ++L
Sbjct 68 KLALKKDR-ESMGHRYIEVFKSHRTEMDWVLKHSGPNSADSANDG-----------FVRL 115
Query 131 RGLPWTCTEMNVVNFFKSM-VLRLGLTL 157
RGLP+ CT+ +V FF + ++ G+TL
Sbjct 116 RGLPFGCTKEEIVQFFSGLEIVPNGITL 143
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 49/80 (61%), Gaps = 2/80 (2%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVRM 76
+RLRGLP+ ++ I++F + EI + + + V ++TGEA+V + E+ + A+
Sbjct 113 VRLRGLPFGCTKEEIVQFFSGL-EIVPNGITLPVDPEGKITGEAFVQFASQELAEKALGK 171
Query 77 LHGRMMGARWIEVFRSSAED 96
H +G R+IEVF+SS E+
Sbjct 172 -HKERIGHRYIEVFKSSQEE 190
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 41/81 (50%), Gaps = 4/81 (4%)
Query 13 VGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDM 72
G + +RGLP+ E+ I F P+ +R V + +G + RVTGEA V T E
Sbjct 287 TGHCVHMRGLPYKATENDIYNFFSPLNPVR---VHIEIGPDGRVTGEADVEFATHEEAVA 343
Query 73 AVRMLHGRMMGARWIEVFRSS 93
A+ M R+IE+F +S
Sbjct 344 AMSKDRANMQ-HRYIELFLNS 363
> hsa:3187 HNRNPH1, DKFZp686A15170, HNRPH, HNRPH1, hnRNPH; heterogeneous
nuclear ribonucleoprotein H1 (H); K12898 heterogeneous
nuclear ribonucleoprotein F/H
Length=449
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 37/148 (25%), Positives = 71/148 (47%), Gaps = 14/148 (9%)
Query 12 GVGTHIRLRGLPWDVNEDAIIRFVKPV-VEIRESDVCVCVGLNKRVTGEAYVNVHTTEMR 70
G G +++RGLPW + D + RF ++ + R +GEA+V + + +
Sbjct 8 GEGFVVKVRGLPWSCSADEVQRFFSDCKIQNGAQGIRFIYTREGRPSGEAFVELESEDEV 67
Query 71 DMAVRMLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGRDADVKVLNLTVLKL 130
+A++ MG R++EVF+S+ + + + + DG ++L
Sbjct 68 KLALKKDR-ETMGHRYVEVFKSNNVEMDWVLKHTGPNSPDTANDG-----------FVRL 115
Query 131 RGLPWTCTEMNVVNFFKSM-VLRLGLTL 157
RGLP+ C++ +V FF + ++ G+TL
Sbjct 116 RGLPFGCSKEEIVQFFSGLEIVPNGITL 143
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/91 (32%), Positives = 53/91 (58%), Gaps = 8/91 (8%)
Query 3 PTPASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYV 62
P A++G +RLRGLP+ +++ I++F + EI + + + V R TGEA+V
Sbjct 105 PDTANDGF------VRLRGLPFGCSKEEIVQFFSGL-EIVPNGITLPVDFQGRSTGEAFV 157
Query 63 NVHTTEMRDMAVRMLHGRMMGARWIEVFRSS 93
+ E+ + A++ H +G R+IE+F+SS
Sbjct 158 QFASQEIAEKALKK-HKERIGHRYIEIFKSS 187
Score = 42.4 bits (98), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 28/82 (34%), Positives = 42/82 (51%), Gaps = 6/82 (7%)
Query 13 VGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDM 72
G + +RGLP+ E+ I F P+ +R V + +G + RVTGEA V T E D
Sbjct 287 TGHCVHMRGLPYRATENDIYNFFSPLNPVR---VHIEIGPDGRVTGEADVEFATHE--DA 341
Query 73 AVRMLHGRM-MGARWIEVFRSS 93
M + M R++E+F +S
Sbjct 342 VAAMSKDKANMQHRYVELFLNS 363
> mmu:59013 Hnrnph1, AI642080, E430005G16Rik, Hnrnph, Hnrph1;
heterogeneous nuclear ribonucleoprotein H1; K12898 heterogeneous
nuclear ribonucleoprotein F/H
Length=449
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 37/148 (25%), Positives = 71/148 (47%), Gaps = 14/148 (9%)
Query 12 GVGTHIRLRGLPWDVNEDAIIRFVKPV-VEIRESDVCVCVGLNKRVTGEAYVNVHTTEMR 70
G G +++RGLPW + D + RF ++ + R +GEA+V + + +
Sbjct 8 GEGFVVKVRGLPWSCSADEVQRFFSDCKIQNGAQGIRFIYTREGRPSGEAFVELESEDEV 67
Query 71 DMAVRMLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGRDADVKVLNLTVLKL 130
+A++ MG R++EVF+S+ + + + + DG ++L
Sbjct 68 KLALKKDR-ETMGHRYVEVFKSNNVEMDWVLKHTGPNSPDTANDG-----------FVRL 115
Query 131 RGLPWTCTEMNVVNFFKSM-VLRLGLTL 157
RGLP+ C++ +V FF + ++ G+TL
Sbjct 116 RGLPFGCSKEEIVQFFSGLEIVPNGITL 143
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/91 (32%), Positives = 53/91 (58%), Gaps = 8/91 (8%)
Query 3 PTPASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYV 62
P A++G +RLRGLP+ +++ I++F + EI + + + V R TGEA+V
Sbjct 105 PDTANDGF------VRLRGLPFGCSKEEIVQFFSGL-EIVPNGITLPVDFQGRSTGEAFV 157
Query 63 NVHTTEMRDMAVRMLHGRMMGARWIEVFRSS 93
+ E+ + A++ H +G R+IE+F+SS
Sbjct 158 QFASQEIAEKALKK-HKERIGHRYIEIFKSS 187
Score = 42.4 bits (98), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 28/82 (34%), Positives = 42/82 (51%), Gaps = 6/82 (7%)
Query 13 VGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDM 72
G + +RGLP+ E+ I F P+ +R V + +G + RVTGEA V T E D
Sbjct 287 TGHCVHMRGLPYRATENDIYNFFSPLNPVR---VHIEIGPDGRVTGEADVEFATHE--DA 341
Query 73 AVRMLHGRM-MGARWIEVFRSS 93
M + M R++E+F +S
Sbjct 342 VAAMSKDKANMQHRYVELFLNS 363
> xla:432071 hnrnph1-a, MGC78776, hnrnph, hnrnph2, hnrph, hnrph1;
heterogeneous nuclear ribonucleoprotein H1 (H); K12898 heterogeneous
nuclear ribonucleoprotein F/H
Length=441
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/91 (32%), Positives = 53/91 (58%), Gaps = 8/91 (8%)
Query 3 PTPASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYV 62
P A++G +RLRGLP+ +++ I++F + EI + + + V R TGEA+V
Sbjct 104 PDTANDGF------VRLRGLPFGCSKEEIVQFFSGL-EIVPNGITLPVDFQGRSTGEAFV 156
Query 63 NVHTTEMRDMAVRMLHGRMMGARWIEVFRSS 93
+ E+ + A++ H +G R+IE+F+SS
Sbjct 157 QFASQEIAEKALKK-HKERIGHRYIEIFKSS 186
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 38/143 (26%), Positives = 68/143 (47%), Gaps = 14/143 (9%)
Query 17 IRLRGLPWDVNEDAIIRFV-KPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVR 75
+++RGLPW + D I F + + S V R +GEA+V T + +AV+
Sbjct 12 VKVRGLPWSCSHDEIENFFSESKIANGLSGVHFIYTREGRPSGEAFVEFETEDDLQLAVK 71
Query 76 MLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGRDADVKVLNLTVLKLRGLPW 135
M R++EVF+S++ + + + + DG ++LRGLP+
Sbjct 72 KDRA-TMAHRYVEVFKSNSVEMDWVLKHTGPNSPDTANDG-----------FVRLRGLPF 119
Query 136 TCTEMNVVNFFKSM-VLRLGLTL 157
C++ +V FF + ++ G+TL
Sbjct 120 GCSKEEIVQFFSGLEIVPNGITL 142
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 41/82 (50%), Gaps = 6/82 (7%)
Query 13 VGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDM 72
G + +RGLP+ E I F P+ +R V + +G + RVTGEA V + E D
Sbjct 275 TGHCVHMRGLPYRATETDIYTFFSPLNPVR---VHIEIGADGRVTGEADVEFASHE--DA 329
Query 73 AVRMLHGRM-MGARWIEVFRSS 93
M + M R++E+F +S
Sbjct 330 VAAMSKDKANMQHRYVELFLNS 351
Score = 35.8 bits (81), Expect = 0.059, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 127 VLKLRGLPWTCTEMNVVNFFKSMVLRLGLT 156
V+K+RGLPW+C+ + NFF + GL+
Sbjct 11 VVKVRGLPWSCSHDEIENFFSESKIANGLS 40
> ath:AT3G20890 RNA binding / nucleic acid binding / nucleotide
binding; K12898 heterogeneous nuclear ribonucleoprotein F/H
Length=278
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 35/88 (39%), Positives = 48/88 (54%), Gaps = 4/88 (4%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVRM 76
+RLRGLP+ ++ I+ F K E+ E V V V R TGEA+V E D M
Sbjct 192 LRLRGLPFSAGKEDILDFFKDF-ELSEDFVHVTVNGEGRPTGEAFVEFRNAE--DSRAAM 248
Query 77 LHGR-MMGARWIEVFRSSAEDFERAQQR 103
+ R +G+R+IE+F SS E+ E A R
Sbjct 249 VKDRKTLGSRYIELFPSSVEELEEALSR 276
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 44/180 (24%), Positives = 78/180 (43%), Gaps = 56/180 (31%)
Query 18 RLRGLPWDVNEDAIIRFVK--PVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVR 75
RLRGLP+D E ++ F VV++ + V N +VTGEA+ + D A++
Sbjct 46 RLRGLPFDCAELDVVEFFHGLDVVDV------LFVHRNNKVTGEAFCVLGYPLQVDFALQ 99
Query 76 MLHGRMMGARWIEVFRSSAEDFERA----------------------------------- 100
+ + MG R++EVFRS+ +++ +A
Sbjct 100 K-NRQNMGRRYVEVFRSTKQEYYKAIANEVAESRVHGMASGGGGGLGGGNGSGGGGGGGG 158
Query 101 ---------QQRRVAMLSSDSRDGRDADVKVLNLTVLKLRGLPWTCTEMNVVNFFKSMVL 151
RR + S DG++ + + +L+LRGLP++ + ++++FFK L
Sbjct 159 GGGRISGGSSPRRHVQRARSSDDGKE---DIEHTGILRLRGLPFSAGKEDILDFFKDFEL 215
> mmu:231413 Grsf1, B130010H02, BB232551, C80280, D5Wsu31e; G-rich
RNA sequence binding factor 1
Length=362
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 38/135 (28%), Positives = 69/135 (51%), Gaps = 15/135 (11%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNK--RVTGEAYVNVHTTEMRDMAV 74
IR +GLPW + ++ F IR S+ + LN+ + G+A + + + + A+
Sbjct 34 IRAQGLPWSCTVEDVLNFFSDC-RIRNSENGIHFLLNRDGKRRGDALIEMESEQDVQKAL 92
Query 75 RMLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGRDADVKVLNLTVLKLRGLP 134
H MG R++EV+ + ED + A + + + S VL+ V++LRGLP
Sbjct 93 EK-HRMYMGQRYVEVYEINNEDVD-ALMKSLQVKPS----------PVLSDGVVRLRGLP 140
Query 135 WTCTEMNVVNFFKSM 149
++C E ++V+FF +
Sbjct 141 YSCNEKDIVDFFAGL 155
Score = 48.9 bits (115), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 34/93 (36%), Positives = 49/93 (52%), Gaps = 14/93 (15%)
Query 3 PTPA-SNGIPGVGTHIRLRGLPWDVNEDAIIRFVK--PVVEIRESDVCVCVGLNKRVTGE 59
P+P S+G+ +RLRGLP+ NE I+ F +V+I V +R TGE
Sbjct 125 PSPVLSDGV------VRLRGLPYSCNEKDIVDFFAGLNIVDI----TFVMDYRGRRKTGE 174
Query 60 AYVNVHTTEMRDMAVRMLHGRMMGARWIEVFRS 92
AYV EM + A+ + H +G R+IE+F S
Sbjct 175 AYVQFEEPEMANQAL-LKHREEIGNRYIEIFPS 206
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 40/78 (51%), Gaps = 6/78 (7%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVRM 76
+ +RGLP+ N II F P+ +R + + + + TGEA V+ T E D M
Sbjct 285 VHMRGLPFQANAQDIINFFAPLKPVR---ITMEYSSSGKATGEADVHFDTHE--DAVAAM 339
Query 77 LHGR-MMGARWIEVFRSS 93
L R + R+IE+F +S
Sbjct 340 LKDRSHVQHRYIELFLNS 357
Score = 33.1 bits (74), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 12/32 (37%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 121 KVLNLTVLKLRGLPWTCTEMNVVNFFKSMVLR 152
+V ++ +++ +GLPW+CT +V+NFF +R
Sbjct 27 EVDDVYLIRAQGLPWSCTVEDVLNFFSDCRIR 58
> xla:446758 hnrnph1-b, MGC130700, MGC80081, hnrnph, hnrnph1,
hnrph, hnrph1, hnrph2; heterogeneous nuclear ribonucleoprotein
H1 (H); K12898 heterogeneous nuclear ribonucleoprotein F/H
Length=456
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/91 (32%), Positives = 53/91 (58%), Gaps = 8/91 (8%)
Query 3 PTPASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYV 62
P A++G +RLRGLP+ +++ I++F + EI + + + V R TGEA+V
Sbjct 104 PDTANDGF------VRLRGLPFGCSKEEIVQFFSGL-EIVPNGITLPVDFQGRSTGEAFV 156
Query 63 NVHTTEMRDMAVRMLHGRMMGARWIEVFRSS 93
+ E+ + A++ H +G R+IE+F+SS
Sbjct 157 QFASQEIAEKALKK-HKERIGHRYIEIFKSS 186
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 39/144 (27%), Positives = 68/144 (47%), Gaps = 16/144 (11%)
Query 17 IRLRGLPWDVNEDAIIRFVKPV-VEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVR 75
+++RGLPW + D I F + S + R +GEA+V T E D+ +
Sbjct 12 VKVRGLPWSCSHDEIENFFSECKIANGLSGIHFIYTREGRPSGEAFVEFETEE--DLKLG 69
Query 76 MLHGR-MMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGRDADVKVLNLTVLKLRGLP 134
+ R MG R++EVF+S+ + + + + DG ++LRGLP
Sbjct 70 LKKDRATMGHRYVEVFKSNNVEMDWVLKHTGPNSPDTANDG-----------FVRLRGLP 118
Query 135 WTCTEMNVVNFFKSM-VLRLGLTL 157
+ C++ +V FF + ++ G+TL
Sbjct 119 FGCSKEEIVQFFSGLEIVPNGITL 142
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 28/82 (34%), Positives = 41/82 (50%), Gaps = 6/82 (7%)
Query 13 VGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDM 72
G + +RGLP+ E I F P+ +R V + +G + RVTGEA V T E D
Sbjct 275 TGHCVHMRGLPYRATETDIYTFFSPLNPVR---VHIEIGADGRVTGEADVEFATHE--DA 329
Query 73 AVRMLHGRM-MGARWIEVFRSS 93
M + M R++E+F +S
Sbjct 330 VAAMSKDKANMQHRYVELFLNS 351
Score = 37.4 bits (85), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 27/50 (54%), Gaps = 10/50 (20%)
Query 107 MLSSDSRDGRDADVKVLNLTVLKLRGLPWTCTEMNVVNFFKSMVLRLGLT 156
M SS+S DG V+K+RGLPW+C+ + NFF + GL+
Sbjct 1 MSSSESSDG----------FVVKVRGLPWSCSHDEIENFFSECKIANGLS 40
> mmu:432467 Hnrnph3, AA693301, AI666703, Hnrph3; heterogeneous
nuclear ribonucleoprotein H3; K12898 heterogeneous nuclear
ribonucleoprotein F/H
Length=346
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 31/91 (34%), Positives = 53/91 (58%), Gaps = 8/91 (8%)
Query 3 PTPASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYV 62
P AS+G +RLRGLP+ +++ I++F + +EI + + + + R TGEA+V
Sbjct 10 PNDASDGT------VRLRGLPFGCSKEEIVQFFQ-GLEIVPNGITLTMDYQGRSTGEAFV 62
Query 63 NVHTTEMRDMAVRMLHGRMMGARWIEVFRSS 93
+ E+ + A+ H +G R+IE+FRSS
Sbjct 63 QFASKEIAENALGK-HKERIGHRYIEIFRSS 92
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 33/89 (37%), Positives = 46/89 (51%), Gaps = 7/89 (7%)
Query 6 ASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVH 65
AS+G G G + +RGLP+ E+ I F P+ IR V + +G + R TGEA V
Sbjct 187 ASSGFHG-GHFVHMRGLPFRATENDIANFFSPLNPIR---VHIDIGADGRATGEADVEFV 242
Query 66 TTEMRDMAVRMLHGR-MMGARWIEVFRSS 93
T E D M + M R+IE+F +S
Sbjct 243 THE--DAVAAMSKDKNNMQHRYIELFLNS 269
Score = 32.3 bits (72), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 27/41 (65%), Gaps = 2/41 (4%)
Query 127 VLKLRGLPWTCTEMNVVNFFKSM-VLRLGLTLSC-WASRWT 165
++LRGLP+ C++ +V FF+ + ++ G+TL+ + R T
Sbjct 17 TVRLRGLPFGCSKEEIVQFFQGLEIVPNGITLTMDYQGRST 57
> hsa:3189 HNRNPH3, 2H9, FLJ34092, HNRPH3; heterogeneous nuclear
ribonucleoprotein H3 (2H9); K12898 heterogeneous nuclear
ribonucleoprotein F/H
Length=346
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 31/91 (34%), Positives = 53/91 (58%), Gaps = 8/91 (8%)
Query 3 PTPASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYV 62
P AS+G +RLRGLP+ +++ I++F + +EI + + + + R TGEA+V
Sbjct 10 PNDASDGT------VRLRGLPFGCSKEEIVQFFQ-GLEIVPNGITLTMDYQGRSTGEAFV 62
Query 63 NVHTTEMRDMAVRMLHGRMMGARWIEVFRSS 93
+ E+ + A+ H +G R+IE+FRSS
Sbjct 63 QFASKEIAENALGK-HKERIGHRYIEIFRSS 92
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 33/89 (37%), Positives = 46/89 (51%), Gaps = 7/89 (7%)
Query 6 ASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVH 65
AS+G G G + +RGLP+ E+ I F P+ IR V + +G + R TGEA V
Sbjct 187 ASSGFHG-GHFVHMRGLPFRATENDIANFFSPLNPIR---VHIDIGADGRATGEADVEFV 242
Query 66 TTEMRDMAVRMLHGR-MMGARWIEVFRSS 93
T E D M + M R+IE+F +S
Sbjct 243 THE--DAVAAMSKDKNNMQHRYIELFLNS 269
Score = 32.0 bits (71), Expect = 0.87, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 27/41 (65%), Gaps = 2/41 (4%)
Query 127 VLKLRGLPWTCTEMNVVNFFKSM-VLRLGLTLSC-WASRWT 165
++LRGLP+ C++ +V FF+ + ++ G+TL+ + R T
Sbjct 17 TVRLRGLPFGCSKEEIVQFFQGLEIVPNGITLTMDYQGRST 57
> dre:387255 rbm19, npo; RNA binding motif protein 19; K14787
multiple RNA-binding domain-containing protein 1
Length=926
Score = 52.8 bits (125), Expect = 5e-07, Method: Composition-based stats.
Identities = 39/134 (29%), Positives = 69/134 (51%), Gaps = 10/134 (7%)
Query 17 IRLRGLPWDVNEDAIIRFVKPV--VEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAV 74
++LRG P++V E + F+ P+ V IR + + R +G YV++ + + A+
Sbjct 291 VKLRGAPFNVKEQQVKEFMMPLKPVAIRFAK-----NSDGRNSGYVYVDLRSEAEVERAL 345
Query 75 RMLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGR--DADVKVLNLTVLKLRG 132
R L MG R+IEVFR++ +R +R M + R+ + + + V L +R
Sbjct 346 R-LDKDYMGGRYIEVFRANNFKNDRRSSKRSEMEKNFVRELKDDEEEEDVAESGRLFIRN 404
Query 133 LPWTCTEMNVVNFF 146
+P+TCTE ++ F
Sbjct 405 MPYTCTEEDLKEVF 418
> bbo:BBOV_III001020 17.m07117; hypothetical protein
Length=837
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 28/90 (31%), Positives = 44/90 (48%), Gaps = 3/90 (3%)
Query 2 LPTPASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAY 61
LP P+ G P + L+GLP+D E I+ ++ + DV + TG+AY
Sbjct 130 LPLPSDPGDPSYDALLYLKGLPYDCVESDILDWLSSYSIV---DVILIKNEEGCFTGDAY 186
Query 62 VNVHTTEMRDMAVRMLHGRMMGARWIEVFR 91
V T RD R + G+ +G R+I ++R
Sbjct 187 VRCSTLSERDRVHREMSGKYLGLRYIPIYR 216
> xla:494658 hnrnph3, hnrph3; heterogeneous nuclear ribonucleoprotein
H3 (2H9); K12898 heterogeneous nuclear ribonucleoprotein
F/H
Length=342
Score = 48.9 bits (115), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 26/77 (33%), Positives = 46/77 (59%), Gaps = 2/77 (2%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVRM 76
+RLRGLP+ +++ I++F + I + + + V R TGEA+V + E+ + A+
Sbjct 36 VRLRGLPFGCSKEEIVQFFSG-LRIVPNGITLTVDYQGRSTGEAFVQFASKEIAENALGK 94
Query 77 LHGRMMGARWIEVFRSS 93
H +G R+IE+F+SS
Sbjct 95 -HKERIGHRYIEIFKSS 110
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 33/91 (36%), Positives = 45/91 (49%), Gaps = 8/91 (8%)
Query 6 ASNGIPGV--GTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVN 63
A +G G G + +RGLP+ +E I F P+ IR V + VG + R TGEA V
Sbjct 200 AGDGSAGFHSGHFVHMRGLPFRASESDIANFFSPLTPIR---VHIDVGADGRATGEADVE 256
Query 64 VHTTEMRDMAVRMLHGR-MMGARWIEVFRSS 93
T E D M + M R+IE+F +S
Sbjct 257 FATHE--DAVAAMSKDKNNMQHRYIELFLNS 285
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 28/86 (32%), Positives = 48/86 (55%), Gaps = 13/86 (15%)
Query 82 MGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGRDADVKVLNLTVLKLRGLPWTCTEMN 141
MG R+IEVF+S+ + E +L +S D DV+ + ++LRGLP+ C++
Sbjct 1 MGHRYIEVFKSNNTEME-------WVLKHNSTD----DVETDSDGTVRLRGLPFGCSKEE 49
Query 142 VVNFFKSM-VLRLGLTLSC-WASRWT 165
+V FF + ++ G+TL+ + R T
Sbjct 50 IVQFFSGLRIVPNGITLTVDYQGRST 75
> hsa:2926 GRSF1, FLJ13125; G-rich RNA sequence binding factor
1
Length=318
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/78 (38%), Positives = 42/78 (53%), Gaps = 7/78 (8%)
Query 17 IRLRGLPWDVNEDAIIRFVK--PVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAV 74
+RLRGLP+ NE I+ F +V+I V +R TGEAYV EM + A+
Sbjct 90 VRLRGLPYSCNEKDIVDFFAGLNIVDI----TFVMDYRGRRKTGEAYVQFEEPEMANQAL 145
Query 75 RMLHGRMMGARWIEVFRS 92
+ H +G R+IE+F S
Sbjct 146 -LKHREEIGNRYIEIFPS 162
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 24/72 (33%), Positives = 41/72 (56%), Gaps = 11/72 (15%)
Query 78 HGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGRDADVKVLNLTVLKLRGLPWTC 137
H MG R++EV+ + ED + A + + + SS V+N V++LRGLP++C
Sbjct 51 HRMYMGQRYVEVYEINNEDVD-ALMKSLQVKSS----------PVVNDGVVRLRGLPYSC 99
Query 138 TEMNVVNFFKSM 149
E ++V+FF +
Sbjct 100 NEKDIVDFFAGL 111
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 30/93 (32%), Positives = 44/93 (47%), Gaps = 6/93 (6%)
Query 2 LPTPASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAY 61
LP A G + +RGLP+ N II F P+ +R + + + + TGEA
Sbjct 226 LPEAADFGTTSSLHFVHMRGLPFQANAQDIINFFAPLKPVR---ITMEYSSSGKATGEAD 282
Query 62 VNVHTTEMRDMAVRMLHGR-MMGARWIEVFRSS 93
V+ T E D ML R + R+IE+F +S
Sbjct 283 VHFETHE--DAVAAMLKDRSHVHHRYIELFLNS 313
> cel:W02D3.11 hrpf-1; HnRNP F homolog family member (hrpf-1)
Length=549
Score = 47.0 bits (110), Expect = 3e-05, Method: Composition-based stats.
Identities = 32/95 (33%), Positives = 49/95 (51%), Gaps = 5/95 (5%)
Query 5 PASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNV 64
P S+G + +RLRGLP+ V I F+ P+ +R+ + + R GEAYV
Sbjct 116 PRSSGPDSI---VRLRGLPFSVTSRDISDFLAPLPIVRDG-ILLPDQQRARPGGEAYVCF 171
Query 65 HTTEMRDMAVRMLHGRMMGARWIEVFRSSAEDFER 99
T E +A + H + +G R+IEVF ++ D R
Sbjct 172 ETMESVQIA-KQRHMKNIGHRYIEVFEATHRDLSR 205
Score = 34.7 bits (78), Expect = 0.15, Method: Composition-based stats.
Identities = 36/158 (22%), Positives = 64/158 (40%), Gaps = 26/158 (16%)
Query 17 IRLRGLPWDVNEDAIIRFVK----PVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDM 72
++ RGLPW+ E + F +EI N R +G+A V E +
Sbjct 7 VQCRGLPWEATEQELRDFFGNNGIESLEIPRR--------NGRTSGDAKVVFTNEEDYNN 58
Query 73 AVRMLHGRMMGARWIEVFRSSAEDFERAQQ------------RRVAMLSSDSRDGRDADV 120
A++ +G+R+IEVF + R + R D R
Sbjct 59 ALKK-DREHLGSRYIEVFPAGGAPTRRGDRGERGDRGDRDHYRSRGAPPRDRYSDRGGPR 117
Query 121 KVLNLTVLKLRGLPWTCTEMNVVNFFKSM-VLRLGLTL 157
++++LRGLP++ T ++ +F + ++R G+ L
Sbjct 118 SSGPDSIVRLRGLPFSVTSRDISDFLAPLPIVRDGILL 155
> tpv:TP03_0737 hypothetical protein
Length=968
Score = 45.8 bits (107), Expect = 6e-05, Method: Composition-based stats.
Identities = 28/101 (27%), Positives = 47/101 (46%), Gaps = 13/101 (12%)
Query 2 LPTPASNGIPGVGTHIRLRGLPWDVNEDAIIRFVK-----PVVEIRESDVCVCVGLNKRV 56
LPTP P + L+G+P+ E + ++K V+ I+ N
Sbjct 166 LPTPPGIYSPSYDALLYLKGIPFKATEKDVFDWLKNYDIVSVIFIKNE--------NGFF 217
Query 57 TGEAYVNVHTTEMRDMAVRMLHGRMMGARWIEVFRSSAEDF 97
TG+AYV ++RD + + + +GAR+I+VFR S +
Sbjct 218 TGDAYVRCVNIQVRDKVAKEMENKRIGARYIQVFRVSENAY 258
> mmu:74111 Rbm19, 1200009A02Rik, AV302714, KIAA0682, NPO; RNA
binding motif protein 19; K14787 multiple RNA-binding domain-containing
protein 1
Length=952
Score = 44.7 bits (104), Expect = 1e-04, Method: Composition-based stats.
Identities = 35/146 (23%), Positives = 64/146 (43%), Gaps = 8/146 (5%)
Query 5 PASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPV--VEIRESDVCVCVGLNKRVTGEAYV 62
P S P ++LRG P++V E +I F+ P+ V IR + + TG +V
Sbjct 283 PVSQKEPTTPYTVKLRGAPFNVTEKNVIEFLAPLKPVAIR-----IVRNAHGNKTGYVFV 337
Query 63 NVHTTEMRDMAVRMLHGRMMGARWIEVFRSSAEDFERAQQRRVAMLSSDSRDGRDADVKV 122
++ + E A++ + MG R+IEVFR R + + + + +
Sbjct 338 DLSSEEEVKKALK-CNRDYMGGRYIEVFREKQAPTARGPPKSTTPWQGRTLGENEEEEDL 396
Query 123 LNLTVLKLRGLPWTCTEMNVVNFFKS 148
+ L +R L +T +E ++ F +
Sbjct 397 ADSGRLFVRNLSYTSSEEDLEKLFSA 422
> xla:494769 rbm19; RNA binding motif protein 19; K14787 multiple
RNA-binding domain-containing protein 1
Length=920
Score = 44.7 bits (104), Expect = 2e-04, Method: Composition-based stats.
Identities = 41/149 (27%), Positives = 65/149 (43%), Gaps = 30/149 (20%)
Query 11 PGVGTHIRLRGLPWDVNEDAIIRFVKPV--VEIRESDVCVCVGLNKRVTGEAYVNVHTTE 68
P ++LRG P++V E + F+ P+ V IR + TG +V++ + E
Sbjct 275 PTTSYTVKLRGAPFNVTEQNVKEFLVPLKPVAIR-----IARNTYGNKTGYVFVDLSSEE 329
Query 69 MRDMAVRMLHGRMMGARWIEVFR----------SSAEDFERAQQRRVAMLSSD-SRDGRD 117
A++ + MG R+IEVFR S AE Q+ + + D S GR
Sbjct 330 EVQKALKR-NKDYMGGRYIEVFRDNYTKSPSVQSKAESRPWEQRDKQELQQEDLSESGR- 387
Query 118 ADVKVLNLTVLKLRGLPWTCTEMNVVNFF 146
L +R LP++CTE ++ F
Sbjct 388 ----------LFVRNLPYSCTEDDLDKLF 406
> hsa:10137 RBM12, HRIHFB2091, KIAA0765, SWAN; RNA binding motif
protein 12
Length=932
Score = 44.3 bits (103), Expect = 2e-04, Method: Composition-based stats.
Identities = 36/142 (25%), Positives = 68/142 (47%), Gaps = 9/142 (6%)
Query 14 GTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMA 73
G + L+GLP++ +I F K ++I E + + G N + TGE +V E A
Sbjct 429 GFCVYLKGLPFEAENKHVIDFFKK-LDIVEDSIYIAYGPNGKATGEGFVEFR-NEADYKA 486
Query 74 VRMLHGRMMGARWIEVFRSSAED-FERAQ--QRRVAMLSSDSRD---GRDADVKVLNLTV 127
H + MG R+I+V + + E+ ++R+ S D R+ + DV +
Sbjct 487 ALCRHKQYMGNRFIQVHPITKKGMLEKIDMIRKRLQNFSYDQREMILNPEGDVNSAKVCA 546
Query 128 LKLRGLPWTCTEMNVVNFFKSM 149
+ +P++ T+M+V+ F + +
Sbjct 547 -HITNIPFSITKMDVLQFLEGI 567
Score = 32.0 bits (71), Expect = 0.91, Method: Composition-based stats.
Identities = 27/86 (31%), Positives = 47/86 (54%), Gaps = 6/86 (6%)
Query 6 ASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKR--VTGEAYVN 63
+S+G PG T I+++ +P+ V+ D I+ F I S VC+ N++ TGEA V
Sbjct 848 SSSGKPG-PTVIKVQNMPFTVSIDEILDFFYGYQVIPGS---VCLKYNEKGMPTGEAMVA 903
Query 64 VHTTEMRDMAVRMLHGRMMGARWIEV 89
+ + AV L+ R +G+R +++
Sbjct 904 FESRDEATAAVIDLNDRPIGSRKVKL 929
> dre:445379 rbm12, zgc:193560, zgc:193570; RNA binding motif
protein 12
Length=876
Score = 43.1 bits (100), Expect = 5e-04, Method: Composition-based stats.
Identities = 34/136 (25%), Positives = 60/136 (44%), Gaps = 3/136 (2%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVRM 76
+ L+GLP++ I F K ++I E + + G N R TGE +V EM A
Sbjct 426 VYLKGLPYEAENKQIFEFFKN-LDIVEDSIYIAYGPNGRATGEGFVEFR-NEMDYKAALG 483
Query 77 LHGRMMGARWIEVFRSSAED-FERAQQRRVAMLSSDSRDGRDADVKVLNLTVLKLRGLPW 135
H + MG+R+I+V + ++ +E+ R M S + + + +P+
Sbjct 484 CHMQYMGSRFIQVHPITKKNMYEKIDAIRKRMQGSQGDQKSSSGGGKSAKSCAHITNIPY 543
Query 136 TCTEMNVVNFFKSMVL 151
T+ +V F + L
Sbjct 544 NVTKKDVRLFLDGIEL 559
Score = 31.2 bits (69), Expect = 1.5, Method: Composition-based stats.
Identities = 25/89 (28%), Positives = 41/89 (46%), Gaps = 4/89 (4%)
Query 1 PLPTPASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEA 60
P P N P T ++++ +P+ V D II F ++ VC+ TGEA
Sbjct 789 PSPGGPGNSRP---TIVKIQNMPFTVTVDEIIDFFYGY-QVLPGSVCLQFSDKGLPTGEA 844
Query 61 YVNVHTTEMRDMAVRMLHGRMMGARWIEV 89
V + + AV L+ R +GAR +++
Sbjct 845 MVAFDSHDEAMAAVMDLNDRPIGARKVKI 873
> hsa:9904 RBM19, DKFZp586F1023, KIAA0682; RNA binding motif protein
19; K14787 multiple RNA-binding domain-containing protein
1
Length=960
Score = 42.7 bits (99), Expect = 6e-04, Method: Composition-based stats.
Identities = 38/152 (25%), Positives = 69/152 (45%), Gaps = 23/152 (15%)
Query 5 PASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPV--VEIRESDVCVCVGLNKRVTGEAYV 62
PA+ P ++LRG P++V E ++ F+ P+ V IR + + TG +V
Sbjct 284 PANQKEPTTCHTVKLRGAPFNVTEKNVMEFLAPLKPVAIR-----IVRNAHGNKTGYIFV 338
Query 63 NVHTTEMRDMAVRMLHGRMMGARWIEVFRS--------SAEDFERAQQRRVAMLSSDSRD 114
+ E A++ + MG R+IEVFR + ++ ++ Q R+ + + D
Sbjct 339 DFSNEEEVKQALK-CNREYMGGRYIEVFREKNVPTTKGAPKNTTKSWQGRILGENEEEED 397
Query 115 GRDADVKVLNLTVLKLRGLPWTCTEMNVVNFF 146
++ L +R LP+T TE ++ F
Sbjct 398 LAESG-------RLFVRNLPYTSTEEDLEKLF 422
> mmu:75710 Rbm12, 5730420G12Rik, 9430070C08Rik, AI852903, MGC30712,
MGC38279, SWAN, mKIAA0765; RNA binding motif protein
12
Length=992
Score = 42.4 bits (98), Expect = 6e-04, Method: Composition-based stats.
Identities = 36/145 (24%), Positives = 68/145 (46%), Gaps = 15/145 (10%)
Query 14 GTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMA 73
G + L+GLP++ +I F K ++I E + + G N + TGE +V A
Sbjct 429 GFCVYLKGLPFEAENKHVIDFFKK-LDIVEDSIYIAYGPNGKATGEGFVEFRNDADYKAA 487
Query 74 VRMLHGRMMGARWIEVFRSSAED-FERAQ--QRRVAMLSSDSR------DGRDADVKVLN 124
+ H + MG R+I+V + + E+ ++R+ S D R +G + KV
Sbjct 488 L-CRHKQYMGNRFIQVHPITKKGMLEKIDMIRKRLQNFSYDQRELVLNPEGEVSSAKV-- 544
Query 125 LTVLKLRGLPWTCTEMNVVNFFKSM 149
+ +P++ T+M+V+ F + +
Sbjct 545 --CAHITNIPFSITKMDVLQFLEGI 567
Score = 33.1 bits (74), Expect = 0.42, Method: Composition-based stats.
Identities = 28/86 (32%), Positives = 47/86 (54%), Gaps = 6/86 (6%)
Query 6 ASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKR--VTGEAYVN 63
AS+G PG T I+++ +P+ V+ D I+ F I S VC+ N++ TGEA V
Sbjct 908 ASSGKPG-PTIIKVQNMPFTVSIDEILDFFYGYQVIPGS---VCLKYNEKGMPTGEAMVA 963
Query 64 VHTTEMRDMAVRMLHGRMMGARWIEV 89
+ + AV L+ R +G+R +++
Sbjct 964 FESRDEATAAVIDLNDRPIGSRKVKL 989
> pfa:PF10_0235 RNA binding protein, putative
Length=160
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/77 (24%), Positives = 42/77 (54%), Gaps = 1/77 (1%)
Query 17 IRLRGLPWDVNEDAIIRFVKPVVEIRES-DVCVCVGLNKRVTGEAYVNVHTTEMRDMAVR 75
++LRGLP+D +E+ I F + +++ + + G+ + TG AYV E A +
Sbjct 57 LKLRGLPFDASEEEIKNFFRDFQLTKQAYPIHIIKGIKNKPTGHAYVYFDDEEEARNACQ 116
Query 76 MLHGRMMGARWIEVFRS 92
++ + + R++E+++
Sbjct 117 AMNRKYIRDRFVEIYQD 133
Score = 35.0 bits (79), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 14/29 (48%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 123 LNLTVLKLRGLPWTCTEMNVVNFFKSMVL 151
+NL LKLRGLP+ +E + NFF+ L
Sbjct 52 INLPRLKLRGLPFDASEEEIKNFFRDFQL 80
> dre:678601 MGC136953; zgc:136953
Length=209
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 26/82 (31%), Positives = 40/82 (48%), Gaps = 8/82 (9%)
Query 14 GTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNV--HTTEMRD 71
G + +RGLP+ E + F P+ +R V + +G N + TGEA V H +
Sbjct 102 GHFVHMRGLPFRATESDVAHFFGPLTPVR---VHIDMGPNGKSTGEADVEFRSHEDAVSA 158
Query 72 MAVRMLHGRMMGARWIEVFRSS 93
M+ H M R+IE+F +S
Sbjct 159 MSKDKNH---MQHRYIELFLNS 177
> xla:379683 rbm12, MGC68861, SWAN; RNA binding motif protein
12
Length=877
Score = 38.5 bits (88), Expect = 0.009, Method: Composition-based stats.
Identities = 25/89 (28%), Positives = 41/89 (46%), Gaps = 2/89 (2%)
Query 1 PLPTPASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEA 60
P P S G + L+GLP++ +I F K + I E + + G N + TGE
Sbjct 416 PQSRPRSRSPHEHGFCVYLKGLPYEAENKHVIDFFKK-LNIVEDSIYIAYGSNGKATGEG 474
Query 61 YVNVHTTEMRDMAVRMLHGRMMGARWIEV 89
++ E A+ H + MG R+++V
Sbjct 475 FLEFRNEEDYKSAL-CRHKQYMGNRFVQV 502
> pfa:PFL2130w conserved Plasmodium protein
Length=1335
Score = 38.1 bits (87), Expect = 0.015, Method: Composition-based stats.
Identities = 24/97 (24%), Positives = 44/97 (45%), Gaps = 1/97 (1%)
Query 2 LPTPASNGIPGVGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAY 61
LP P + G I +GLP+ V+ II F KP +I E + G+
Sbjct 172 LPLPVEKNMEGYNCLILCKGLPFHVDNSQIIEFFKP-YKIMEKYIIFMRDKKGHFFGDIL 230
Query 62 VNVHTTEMRDMAVRMLHGRMMGARWIEVFRSSAEDFE 98
V E + +A++ + + + R+I+++ + E +E
Sbjct 231 VRFINKEQKYLALKNKNYKFLLHRYIQLYNVNEEHYE 267
> dre:100330550 heterogeneous nuclear ribonucleoprotein H3-like
Length=166
Score = 36.6 bits (83), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 18/55 (32%), Positives = 28/55 (50%), Gaps = 3/55 (5%)
Query 14 GTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTE 68
G + +RGLP+ E + F P+ +R V + +G N + TGEA V + E
Sbjct 102 GHFVHMRGLPFRATESDVAHFFGPLTPVR---VHIDMGPNGKSTGEADVEFRSHE 153
> sce:YGL044C RNA15; Cleavage and polyadenylation factor I (CF
I) component involved in cleavage and polyadenylation of mRNA
3' ends; interacts with the A-rich polyadenylation signal
in complex with Rna14p and Hrp1p; K14407 cleavage stimulation
factor subunit 2
Length=296
Score = 34.7 bits (78), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 39/83 (46%), Gaps = 7/83 (8%)
Query 17 IRLRGLPWDVNEDAIIRF---VKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDMA 73
+ L +P+D E+ I+ V PV+ ++ + R G A++ E A
Sbjct 20 VYLGSIPYDQTEEQILDLCSNVGPVINLK----MMFDPQTGRSKGYAFIEFRDLESSASA 75
Query 74 VRMLHGRMMGARWIEVFRSSAED 96
VR L+G +G+R+++ SS D
Sbjct 76 VRNLNGYQLGSRFLKCGYSSNSD 98
> ath:AT4G20030 RNA recognition motif (RRM)-containing protein
Length=152
Score = 34.3 bits (77), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 21/88 (23%), Positives = 41/88 (46%), Gaps = 3/88 (3%)
Query 13 VGTHIRLRGLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEAYVNVHTTEMRDM 72
+ + I +R LP+ +ED + R EI E + + KR G A++ + + +
Sbjct 38 LASKIMVRNLPFSTSEDFLKREFSAFGEIAEVKLIKDEAM-KRSKGYAFIQFTSQDDAFL 96
Query 73 AVRMLHGRMMGAR--WIEVFRSSAEDFE 98
A+ + RM R +I++ + DF+
Sbjct 97 AIETMDRRMYNGRMIYIDIAKPGKRDFQ 124
> dre:100330634 RNA binding motif protein 12-like
Length=675
Score = 32.3 bits (72), Expect = 0.85, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 110 SDSRDGRDADVKVLNLTVLKLRGLPWTCTEMNVVNFFKSM 149
+++R RD D LKL G+P++ T+ NV NFF +
Sbjct 140 NEARRQRDGDAPERAEVYLKLTGMPFSATKDNVHNFFAGL 179
> bbo:BBOV_IV007810 23.m06352; U2 splicing factor subunit; K12836
splicing factor U2AF 35 kDa subunit
Length=251
Score = 31.6 bits (70), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 17/61 (27%), Positives = 26/61 (42%), Gaps = 0/61 (0%)
Query 44 SDVCVCVGLNKRVTGEAYVNVHTTEMRDMAVRMLHGRMMGARWIEVFRSSAEDFERAQQR 103
D+ VC + + G YV A+ ML GR G + I+ + DF A+ R
Sbjct 97 EDMVVCDNIGDHIIGNVYVKYRDENSAAHAISMLSGRFYGGKPIQCEYTPVTDFREARCR 156
Query 104 R 104
+
Sbjct 157 Q 157
> ath:AT4G36960 RNA recognition motif (RRM)-containing protein
Length=379
Score = 31.2 bits (69), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 21/92 (22%), Positives = 44/92 (47%), Gaps = 11/92 (11%)
Query 21 GLPWDVNEDAIIRFVKPVVEIRESDVCVCVGLNKRVTGEA----YVNVHTTEMRDMAVRM 76
G+PWD++ D + ++ + D+ C+ + R TG + YV + E A++
Sbjct 9 GIPWDIDSDGLKDYMS-----KFGDLEDCIVMKDRSTGRSRGFGYVTFASAEDAKNALKG 63
Query 77 LHGRMMGARWIEVFRSSAEDFERAQQRRVAML 108
H +G R +EV ++ ++ R ++V +
Sbjct 64 EH--FLGNRILEVKVATPKEEMRQPAKKVTRI 93
Lambda K H
0.324 0.137 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4027755848
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40