bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6548_orf1
Length=85
Score E
Sequences producing significant alignments: (Bits) Value
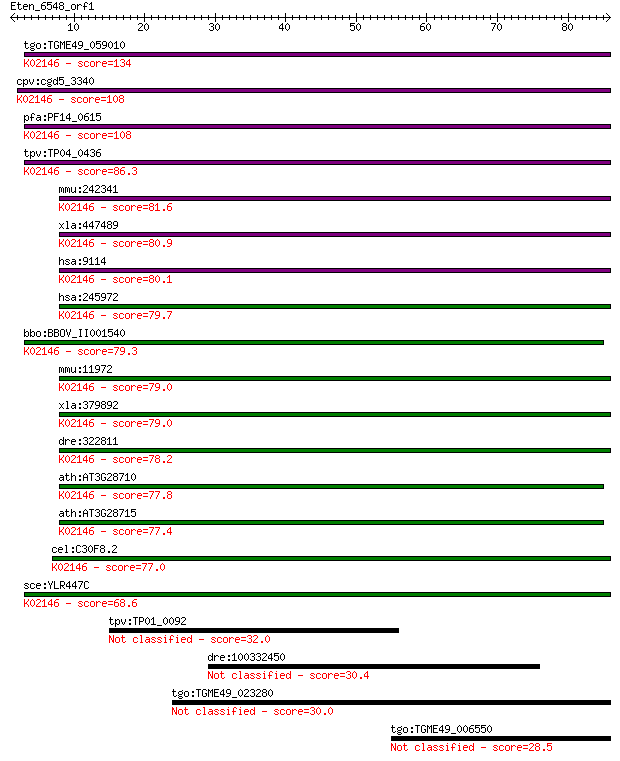
tgo:TGME49_059010 vacuolar ATP synthase subunit D, putative (E... 134 9e-32
cpv:cgd5_3340 vacuolar ATP synthase subunit d ; K02146 V-type ... 108 3e-24
pfa:PF14_0615 ATP synthase (C/AC39) subunit, putative; K02146 ... 108 3e-24
tpv:TP04_0436 vacuolar ATP synthase (C/AC39) subunit (EC:3.6.3... 86.3 2e-17
mmu:242341 Atp6v0d2, 1620401A02Rik, AI324824, V-ATPase; ATPase... 81.6 6e-16
xla:447489 atp6v0d2, MGC81907; ATPase, H+ transporting, lysoso... 80.9 9e-16
hsa:9114 ATP6V0D1, ATP6D, ATP6DV, FLJ43534, P39, VATX, VMA6, V... 80.1 1e-15
hsa:245972 ATP6V0D2, ATP6D2, FLJ38708, VMA6; ATPase, H+ transp... 79.7 2e-15
bbo:BBOV_II001540 18.m06119; vacuolar ATP synthase subunit d; ... 79.3 3e-15
mmu:11972 Atp6v0d1, AI267038, Ac39, Atp6d, P39, VATX, Vma6; AT... 79.0 4e-15
xla:379892 atp6v0d1, MGC53957, atp6d; ATPase, H+ transporting,... 79.0 4e-15
dre:322811 atp6v0d1, fb73h07, wu:fb73h07, zgc:63769; ATPase, H... 78.2 6e-15
ath:AT3G28710 H+-transporting two-sector ATPase, putative; K02... 77.8 7e-15
ath:AT3G28715 H+-transporting two-sector ATPase, putative; K02... 77.4 1e-14
cel:C30F8.2 vha-16; Vacuolar H ATPase family member (vha-16); ... 77.0 1e-14
sce:YLR447C VMA6; Subunit d of the five-subunit V0 integral me... 68.6 5e-12
tpv:TP01_0092 hypothetical protein 32.0 0.48
dre:100332450 retrotransposon protein, putative, Ty3-gypsy sub... 30.4 1.6
tgo:TGME49_023280 hypothetical protein 30.0 1.9
tgo:TGME49_006550 hypothetical protein 28.5 5.3
> tgo:TGME49_059010 vacuolar ATP synthase subunit D, putative
(EC:3.6.3.14); K02146 V-type H+-transporting ATPase subunit
AC39 [EC:3.6.3.14]
Length=396
Score = 134 bits (336), Expect = 9e-32, Method: Compositional matrix adjust.
Identities = 60/83 (72%), Positives = 73/83 (87%), Gaps = 0/83 (0%)
Query 3 MELALFNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPL 62
MEL+ FNV DGYLEGI RGLR +FLT EDYK+L+ AD+LEDLRSALEETDYG FMQDEPL
Sbjct 1 MELSFFNVDDGYLEGICRGLRSAFLTEEDYKKLSAADSLEDLRSALEETDYGPFMQDEPL 60
Query 63 PLAVPTIAQRARSKLTAEFKYLQ 85
PLAVPT++Q+ R K+ +EF+Y++
Sbjct 61 PLAVPTLSQKCREKMASEFRYMR 83
> cpv:cgd5_3340 vacuolar ATP synthase subunit d ; K02146 V-type
H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=412
Score = 108 bits (271), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 50/84 (59%), Positives = 62/84 (73%), Gaps = 0/84 (0%)
Query 2 KMELALFNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEP 61
KMEL FN+ DGYLE +VRG R F+T ++Y + A+TLED+R+ALEETDYG FMQDEP
Sbjct 17 KMELLTFNLKDGYLEAMVRGYRSGFITMDEYHLIGQAETLEDMRTALEETDYGTFMQDEP 76
Query 62 LPLAVPTIAQRARSKLTAEFKYLQ 85
LPL+V I Q+ R K EF+ LQ
Sbjct 77 LPLSVNVITQKCREKFAHEFRMLQ 100
> pfa:PF14_0615 ATP synthase (C/AC39) subunit, putative; K02146
V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=382
Score = 108 bits (271), Expect = 3e-24, Method: Composition-based stats.
Identities = 49/83 (59%), Positives = 63/83 (75%), Gaps = 0/83 (0%)
Query 3 MELALFNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPL 62
MEL +N +GYLE ++RG R SFLT E+YK+L ADTLEDL+S LE+TDYG FM DEP
Sbjct 1 MELCFYNSKNGYLEALLRGFRSSFLTPEEYKKLTEADTLEDLKSVLEDTDYGTFMMDEPS 60
Query 63 PLAVPTIAQRARSKLTAEFKYLQ 85
P+AV TIAQ+ + K+ EF Y++
Sbjct 61 PIAVTTIAQKCKEKMAHEFNYIR 83
> tpv:TP04_0436 vacuolar ATP synthase (C/AC39) subunit (EC:3.6.3.14);
K02146 V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=383
Score = 86.3 bits (212), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 41/83 (49%), Positives = 57/83 (68%), Gaps = 0/83 (0%)
Query 3 MELALFNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPL 62
MEL FNV+ GYLEGIVRG R +FLT DYK++ A++LEDLR+ LE TDY DE
Sbjct 1 MELCTFNVNYGYLEGIVRGYRSTFLTAMDYKKMGVAESLEDLRTVLEATDYTSAFIDEQA 60
Query 63 PLAVPTIAQRARSKLTAEFKYLQ 85
+ I++R + KL ++++YL+
Sbjct 61 QITTKLISKRCKEKLASDYQYLR 83
> mmu:242341 Atp6v0d2, 1620401A02Rik, AI324824, V-ATPase; ATPase,
H+ transporting, lysosomal V0 subunit D2 (EC:3.6.3.14);
K02146 V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=350
Score = 81.6 bits (200), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 39/78 (50%), Positives = 49/78 (62%), Gaps = 0/78 (0%)
Query 8 FNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPLPLAVP 67
FNV GYLEG+VRG + S LT++DY L +TLEDL+ L+ TDYG F+ +E PL V
Sbjct 9 FNVDHGYLEGLVRGCKASLLTQQDYVNLVQCETLEDLKIHLQTTDYGNFLANETNPLTVS 68
Query 68 TIAQRARSKLTAEFKYLQ 85
I R KL EF Y +
Sbjct 69 KIDTEMRKKLCREFDYFR 86
> xla:447489 atp6v0d2, MGC81907; ATPase, H+ transporting, lysosomal
38kDa, V0 subunit d2 (EC:3.6.3.14); K02146 V-type H+-transporting
ATPase subunit AC39 [EC:3.6.3.14]
Length=288
Score = 80.9 bits (198), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 39/78 (50%), Positives = 49/78 (62%), Gaps = 0/78 (0%)
Query 8 FNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPLPLAVP 67
FNV GYLEG+VRG +G L DY LA +TLEDL+ L+ TDYG F+ +E L V
Sbjct 8 FNVDSGYLEGLVRGFKGGILKSTDYLNLAQCETLEDLKLHLQSTDYGSFLANETRQLTVS 67
Query 68 TIAQRARSKLTAEFKYLQ 85
TI Q+ + KL EF Y +
Sbjct 68 TIDQKLKEKLMTEFHYFR 85
> hsa:9114 ATP6V0D1, ATP6D, ATP6DV, FLJ43534, P39, VATX, VMA6,
VPATPD; ATPase, H+ transporting, lysosomal 38kDa, V0 subunit
d1 (EC:3.6.3.14); K02146 V-type H+-transporting ATPase subunit
AC39 [EC:3.6.3.14]
Length=351
Score = 80.1 bits (196), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 36/78 (46%), Positives = 52/78 (66%), Gaps = 0/78 (0%)
Query 8 FNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPLPLAVP 67
FNV +GYLEG+VRGL+ L++ DY L +TLEDL+ L+ TDYG F+ +E PL V
Sbjct 9 FNVDNGYLEGLVRGLKAGVLSQADYLNLVQCETLEDLKLHLQSTDYGNFLANEASPLTVS 68
Query 68 TIAQRARSKLTAEFKYLQ 85
I R + K+ EF++++
Sbjct 69 VIDDRLKEKMVVEFRHMR 86
> hsa:245972 ATP6V0D2, ATP6D2, FLJ38708, VMA6; ATPase, H+ transporting,
lysosomal 38kDa, V0 subunit d2 (EC:3.6.3.14); K02146
V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=350
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 37/78 (47%), Positives = 49/78 (62%), Gaps = 0/78 (0%)
Query 8 FNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPLPLAVP 67
FNV GYLEG+VRG + S LT++DY L +TLEDL+ L+ TDYG F+ + PL V
Sbjct 9 FNVDHGYLEGLVRGCKASLLTQQDYINLVQCETLEDLKIHLQTTDYGNFLANHTNPLTVS 68
Query 68 TIAQRARSKLTAEFKYLQ 85
I R +L EF+Y +
Sbjct 69 KIDTEMRKRLCGEFEYFR 86
> bbo:BBOV_II001540 18.m06119; vacuolar ATP synthase subunit d;
K02146 V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=374
Score = 79.3 bits (194), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 42/82 (51%), Positives = 51/82 (62%), Gaps = 0/82 (0%)
Query 3 MELALFNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPL 62
MELALFN GYLEGIVRG R +FLT DYK++ A+ L+DLR+ LE TDYG E
Sbjct 1 MELALFNAKYGYLEGIVRGYRATFLTPLDYKKMKTAENLDDLRNILEGTDYGTAFYHEQD 60
Query 63 PLAVPTIAQRARSKLTAEFKYL 84
+I +R KL +F YL
Sbjct 61 INNASSIVRRCNEKLANDFAYL 82
> mmu:11972 Atp6v0d1, AI267038, Ac39, Atp6d, P39, VATX, Vma6;
ATPase, H+ transporting, lysosomal V0 subunit D1 (EC:3.6.3.14);
K02146 V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=351
Score = 79.0 bits (193), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 35/78 (44%), Positives = 52/78 (66%), Gaps = 0/78 (0%)
Query 8 FNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPLPLAVP 67
FNV +GYLEG+VRGL+ L++ DY L +TLEDL+ L+ TDYG F+ +E PL V
Sbjct 9 FNVDNGYLEGLVRGLKAGVLSQADYLNLVQCETLEDLKLHLQSTDYGNFLANEASPLTVS 68
Query 68 TIAQRARSKLTAEFKYLQ 85
I + + K+ EF++++
Sbjct 69 VIDDKLKEKMVVEFRHMR 86
> xla:379892 atp6v0d1, MGC53957, atp6d; ATPase, H+ transporting,
lysosomal 38kDa, V0 subunit d1 (EC:3.6.3.14); K02146 V-type
H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=351
Score = 79.0 bits (193), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 35/78 (44%), Positives = 51/78 (65%), Gaps = 0/78 (0%)
Query 8 FNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPLPLAVP 67
FNV GYLEG+VRG + L++ DY L +TLEDL+ L+ TDYG F+ +E PLAV
Sbjct 9 FNVDSGYLEGLVRGFKAGILSQGDYLNLVQCETLEDLKLHLQSTDYGTFLANEASPLAVS 68
Query 68 TIAQRARSKLTAEFKYLQ 85
I + + K+ EF++++
Sbjct 69 VIDDKLKEKMVVEFRHMR 86
> dre:322811 atp6v0d1, fb73h07, wu:fb73h07, zgc:63769; ATPase,
H+ transporting, V0 subunit D isoform 1 (EC:3.6.3.14); K02146
V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=350
Score = 78.2 bits (191), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 34/78 (43%), Positives = 51/78 (65%), Gaps = 0/78 (0%)
Query 8 FNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPLPLAVP 67
FNV +GYLEG+VRG + L++ DY L +TLEDL+ L+ TDYG F+ +E PL V
Sbjct 8 FNVDNGYLEGLVRGFKAGILSQADYLNLVQCETLEDLKLHLQSTDYGSFLANEASPLTVS 67
Query 68 TIAQRARSKLTAEFKYLQ 85
I + + K+ EF++++
Sbjct 68 VIDDKLKEKMVVEFRHMR 85
> ath:AT3G28710 H+-transporting two-sector ATPase, putative; K02146
V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=351
Score = 77.8 bits (190), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 33/77 (42%), Positives = 47/77 (61%), Gaps = 0/77 (0%)
Query 8 FNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPLPLAVP 67
FN+H GYLE IVRG R LT DY L + L+D++ L T YG ++Q+EP PL
Sbjct 9 FNIHGGYLEAIVRGHRAGLLTTADYNNLCQCENLDDIKMHLSATKYGSYLQNEPSPLHTT 68
Query 68 TIAQRARSKLTAEFKYL 84
TI ++ KL ++K++
Sbjct 69 TIVEKCTLKLVDDYKHM 85
> ath:AT3G28715 H+-transporting two-sector ATPase, putative; K02146
V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=351
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 33/77 (42%), Positives = 47/77 (61%), Gaps = 0/77 (0%)
Query 8 FNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPLPLAVP 67
FN+H GYLE IVRG R LT DY L + L+D++ L T YG ++Q+EP PL
Sbjct 9 FNIHGGYLEAIVRGHRAGLLTTADYNNLCQCENLDDIKMHLSATKYGPYLQNEPSPLHTT 68
Query 68 TIAQRARSKLTAEFKYL 84
TI ++ KL ++K++
Sbjct 69 TIVEKCTLKLVDDYKHM 85
> cel:C30F8.2 vha-16; Vacuolar H ATPase family member (vha-16);
K02146 V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=348
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 34/79 (43%), Positives = 50/79 (63%), Gaps = 0/79 (0%)
Query 7 LFNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPLPLAV 66
LFN+ GYLE + RGL+G L + DY L +TLEDL+ ++ TDYG F+ +EP + V
Sbjct 5 LFNIDHGYLEALTRGLKGGLLAQADYANLVQCETLEDLKLHIQSTDYGNFLANEPGAITV 64
Query 67 PTIAQRARSKLTAEFKYLQ 85
I ++ + KL EF +L+
Sbjct 65 QVIDEKLKEKLVTEFTHLR 83
> sce:YLR447C VMA6; Subunit d of the five-subunit V0 integral
membrane domain of vacuolar H+-ATPase (V-ATPase), an electrogenic
proton pump found in the endomembrane system; stabilizes
VO subunits; required for V1 domain assembly on the vacuolar
membrane (EC:3.6.3.14); K02146 V-type H+-transporting ATPase
subunit AC39 [EC:3.6.3.14]
Length=345
Score = 68.6 bits (166), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 35/84 (41%), Positives = 46/84 (54%), Gaps = 1/84 (1%)
Query 3 MELALFNVHDGYLEGIVRGLRGSFLTREDYKRLANADTLEDLRSALEETDYGFFMQD-EP 61
ME FN+ +G++EG+VRG R L+ Y L DTLEDL+ L TDYG F+
Sbjct 1 MEGVYFNIDNGFIEGVVRGYRNGLLSNNQYINLTQCDTLEDLKLQLSSTDYGNFLSSVSS 60
Query 62 LPLAVPTIAQRARSKLTAEFKYLQ 85
L I + A SKL EF Y++
Sbjct 61 ESLTTSLIQEYASSKLYHEFNYIR 84
> tpv:TP01_0092 hypothetical protein
Length=2607
Score = 32.0 bits (71), Expect = 0.48, Method: Composition-based stats.
Identities = 18/44 (40%), Positives = 25/44 (56%), Gaps = 3/44 (6%)
Query 15 LEGIVRGLRGSFLTR---EDYKRLANADTLEDLRSALEETDYGF 55
+ I RGL+ L + +DYKR++N +TL L S EE GF
Sbjct 2535 VRNIARGLKHDLLAKANQKDYKRVSNVNTLVLLGSCTEEFPQGF 2578
> dre:100332450 retrotransposon protein, putative, Ty3-gypsy subclass-like
Length=778
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 18/47 (38%), Positives = 25/47 (53%), Gaps = 1/47 (2%)
Query 29 REDYKRLANADTLEDLRSALEETDYGFFMQDEPLPLAVPTIAQRARS 75
RE +RLA AD LED+R L+E + + P A P + R +S
Sbjct 46 RERSRRLAPAD-LEDVRLHLQELQSSGIISESRSPYASPIVVVRKKS 91
> tgo:TGME49_023280 hypothetical protein
Length=922
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 32/72 (44%), Gaps = 10/72 (13%)
Query 24 GSFLTREDYKRLANADTLEDLRSALEETDYGFFMQDEPLPLAV----------PTIAQRA 73
GS L R + + A ED+RSA E+T + +D L + PT+A R
Sbjct 25 GSLLERLSLHQSSQATLFEDMRSAEEDTAFQRMSEDLSRSLLIFQESGAGHTSPTVASRR 84
Query 74 RSKLTAEFKYLQ 85
KL A + LQ
Sbjct 85 VHKLEASLEELQ 96
> tgo:TGME49_006550 hypothetical protein
Length=4703
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 11/31 (35%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 55 FFMQDEPLPLAVPTIAQRARSKLTAEFKYLQ 85
+++D P P +P +A+ A K+T + YLQ
Sbjct 2395 LWVRDNPTPAEIPLVARGALPKVTVDADYLQ 2425
Lambda K H
0.320 0.138 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2035463248
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40