bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6558_orf2
Length=158
Score E
Sequences producing significant alignments: (Bits) Value
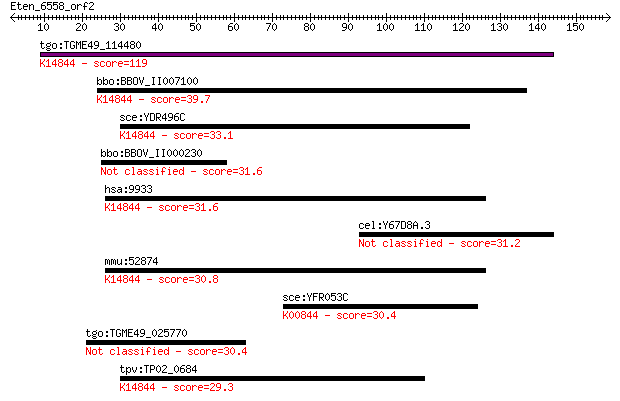
tgo:TGME49_114480 hypothetical protein ; K14844 pumilio homolo... 119 5e-27
bbo:BBOV_II007100 18.m06588; hypothetical protein; K14844 pumi... 39.7 0.005
sce:YDR496C PUF6; Puf6p; K14844 pumilio homology domain family... 33.1 0.37
bbo:BBOV_II000230 18.m06001; serine esterase domain containing... 31.6 1.1
hsa:9933 HLA-HA8, MGC8749, PEN, PUF6, XTP5; KIAA0020; K14844 p... 31.6 1.2
cel:Y67D8A.3 hypothetical protein 31.2 1.5
mmu:52874 D19Bwg1357e, 1110069H02Rik, AA675048, KIAA0020; DNA ... 30.8 1.8
sce:YFR053C HXK1; Hexokinase isoenzyme 1, a cytosolic protein ... 30.4 2.4
tgo:TGME49_025770 protein kinase, putative (EC:2.7.11.25) 30.4 2.4
tpv:TP02_0684 hypothetical protein; K14844 pumilio homology do... 29.3 6.1
> tgo:TGME49_114480 hypothetical protein ; K14844 pumilio homology
domain family member 6
Length=806
Score = 119 bits (297), Expect = 5e-27, Method: Compositional matrix adjust.
Identities = 55/135 (40%), Positives = 84/135 (62%), Gaps = 0/135 (0%)
Query 9 VKSAVNFLEGPLSRKDRHSCTARLAQICLKYAEGDTRKRLWDLLSKDFTVFCECKSFHHV 68
VK +F+ +S DRH CT+R Q LK+ D R++LW +L DF C K+ V
Sbjct 247 VKDLFDFILPRVSEGDRHLCTSRALQALLKFGSKDQRQQLWTILKGDFAELCMGKTMCQV 306
Query 69 CIKMYLYGTTEQKEKLAALLASRRDTAFTKHGAAVWEYIYTSQKTALGQQKLLNSLLLEP 128
+K YLYG +E+++ LL+ ++ F+K GA VWEY+YTS K+A GQQ++LN+++L P
Sbjct 307 AMKFYLYGDRATQEEISQLLSRNKEIFFSKFGARVWEYVYTSTKSAKGQQQMLNAVVLPP 366
Query 129 ALLVAVPNVLQAASF 143
+V P+ +A +F
Sbjct 367 LAMVRHPDYQEAPTF 381
> bbo:BBOV_II007100 18.m06588; hypothetical protein; K14844 pumilio
homology domain family member 6
Length=630
Score = 39.7 bits (91), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 34/121 (28%), Positives = 54/121 (44%), Gaps = 17/121 (14%)
Query 24 DRHSCTA------RLAQICLKYAEGDTRKRLWDLLSKDFTVFCECKSFHHVCIKMYLYGT 77
D +S TA R+ Q CLKY + R +++ L KDF++ IK++ Y
Sbjct 161 DNYSVTANKRHISRVLQACLKYGDTKVRTSIFESLKKDFSLLNLNVHSARFLIKVFHYCN 220
Query 78 TEQKEKLAALLASRRDTA--FTKHGAAVWEYIYTSQKTALGQQKLLNSLLLEPALLVAVP 135
+ K L + ++ F+++G+ V + IY QKL N LE L A+
Sbjct 221 VDAKAFLRQSFFNDKNKVLLFSRYGSLVMDVIY---------QKLRNKEQLEILRLYALS 271
Query 136 N 136
N
Sbjct 272 N 272
> sce:YDR496C PUF6; Puf6p; K14844 pumilio homology domain family
member 6
Length=656
Score = 33.1 bits (74), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 22/93 (23%), Positives = 44/93 (47%), Gaps = 1/93 (1%)
Query 30 ARLAQICLKYAEGDTRKRLWDLLSKDFTVFCECKSFHHVCIKMYLYGTTEQKEKLA-ALL 88
+R+ Q +KY+ D R+++ D L F V ++ +K+ YG+ ++ + L
Sbjct 171 SRIVQTLVKYSSKDRREQIVDALKGKFYVLATSAYGKYLLVKLLHYGSRSSRQTIINELH 230
Query 89 ASRRDTAFTKHGAAVWEYIYTSQKTALGQQKLL 121
S R + GA V E ++ T +Q+++
Sbjct 231 GSLRKLMRHREGAYVVEDLFVLYATHEQRQQMI 263
> bbo:BBOV_II000230 18.m06001; serine esterase domain containing
protein
Length=543
Score = 31.6 bits (70), Expect = 1.1, Method: Composition-based stats.
Identities = 16/36 (44%), Positives = 23/36 (63%), Gaps = 3/36 (8%)
Query 25 RHSCTARLAQICLKYAEGDTRK--RLWDLLSK-DFT 57
RH ++ + +KYAE DT + +WD+LSK DFT
Sbjct 348 RHRVAGKMVKSFIKYAEKDTGEPHSIWDMLSKCDFT 383
> hsa:9933 HLA-HA8, MGC8749, PEN, PUF6, XTP5; KIAA0020; K14844
pumilio homology domain family member 6
Length=648
Score = 31.6 bits (70), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 25/101 (24%), Positives = 44/101 (43%), Gaps = 3/101 (2%)
Query 26 HSCTARLAQICLKYAEGDTRKRLWDLLSKDFTVFCECKSFHHVCIKMYLYGTTEQ-KEKL 84
H T R+ Q ++Y + RK+ ++ L D + K ++ K +YG+ Q E +
Sbjct 177 HDST-RVIQCYIQYGNEEQRKQAFEELRDDLVELSKAKYSRNIVKKFLMYGSKPQIAEII 235
Query 85 AALLASRRDTAFTKHGAAVWEYIYTSQKTALGQQKLLNSLL 125
+ R +A+ EY Y + K L Q+ +L L
Sbjct 236 RSFKGHVRKMLRHAEASAIVEYAY-NDKAILEQRNMLTEEL 275
> cel:Y67D8A.3 hypothetical protein
Length=254
Score = 31.2 bits (69), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 24/51 (47%), Gaps = 0/51 (0%)
Query 93 DTAFTKHGAAVWEYIYTSQKTALGQQKLLNSLLLEPALLVAVPNVLQAASF 143
D A GAA WE I +T + Q L+N + PAL+ + Q SF
Sbjct 177 DIAVDPVGAANWEQITNIVRTTMLNQILMNPTNVSPALISFLLQPTQQPSF 227
> mmu:52874 D19Bwg1357e, 1110069H02Rik, AA675048, KIAA0020; DNA
segment, Chr 19, Brigham & Women's Genetics 1357 expressed;
K14844 pumilio homology domain family member 6
Length=648
Score = 30.8 bits (68), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 25/101 (24%), Positives = 43/101 (42%), Gaps = 3/101 (2%)
Query 26 HSCTARLAQICLKYAEGDTRKRLWDLLSKDFTVFCECKSFHHVCIKMYLYGTTEQ-KEKL 84
H T R+ Q ++Y + RK+ + L D + K ++ K +YG+ Q E +
Sbjct 177 HDST-RVIQCFIQYGNEEQRKQAFQELQGDLVELSKAKYSRNIVKKFLMYGSKPQVAEII 235
Query 85 AALLASRRDTAFTKHGAAVWEYIYTSQKTALGQQKLLNSLL 125
+ R +A+ EY Y + K L Q+ +L L
Sbjct 236 RSFKGHVRKMLRHSEASAIVEYAY-NDKAILEQRNMLTEEL 275
> sce:YFR053C HXK1; Hexokinase isoenzyme 1, a cytosolic protein
that catalyzes phosphorylation of glucose during glucose metabolism;
expression is highest during growth on non-glucose
carbon sources; glucose-induced repression involves the hexokinase
Hxk2p (EC:2.7.1.4 2.7.1.1); K00844 hexokinase [EC:2.7.1.1]
Length=485
Score = 30.4 bits (67), Expect = 2.4, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 6/51 (11%)
Query 73 YLYGTTEQKEKLAALLASRRDTAFTKHGAAVWEYIYTSQKTALGQQKLLNS 123
+ + TT+ K KL D TKH +W +I S K + +Q+LLN+
Sbjct 103 HTFDTTQSKYKLP------HDMRTTKHQEELWSFIADSLKDFMVEQELLNT 147
> tgo:TGME49_025770 protein kinase, putative (EC:2.7.11.25)
Length=1543
Score = 30.4 bits (67), Expect = 2.4, Method: Composition-based stats.
Identities = 13/42 (30%), Positives = 20/42 (47%), Gaps = 0/42 (0%)
Query 21 SRKDRHSCTARLAQICLKYAEGDTRKRLWDLLSKDFTVFCEC 62
S RH+CT + C KY+ R+ WD + ++ C C
Sbjct 581 SSDGRHACTTPASSTCSKYSPFGFRQLSWDSVRSAESLRCAC 622
> tpv:TP02_0684 hypothetical protein; K14844 pumilio homology
domain family member 6
Length=484
Score = 29.3 bits (64), Expect = 6.1, Method: Composition-based stats.
Identities = 20/82 (24%), Positives = 35/82 (42%), Gaps = 2/82 (2%)
Query 30 ARLAQICLKYAEGDTRKRLWDLLSKDFTVFCECKSFHHVCIKMYLYGTTEQKEKLAALL- 88
+R+ Q CLKY + D R + + IK++ Y TE K+ L
Sbjct 113 SRILQACLKYGDSDVRASISQRFRDGLNLTNLNAHSSKFLIKLFHYCNTEVKQFLRTSFF 172
Query 89 -ASRRDTAFTKHGAAVWEYIYT 109
+++ FT+ + V + IY+
Sbjct 173 NEKQKNLIFTRFSSDVMDIIYS 194
Lambda K H
0.320 0.131 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3582056500
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40