bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6709_orf2
Length=51
Score E
Sequences producing significant alignments: (Bits) Value
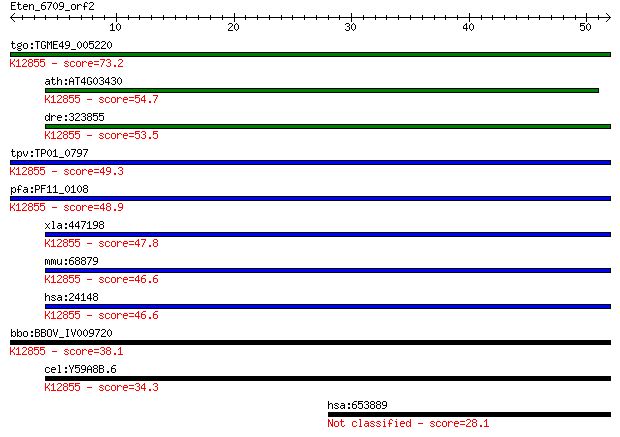
tgo:TGME49_005220 U5 snRNP-associated 102 kDa protein, putativ... 73.2 2e-13
ath:AT4G03430 EMB2770 (EMBRYO DEFECTIVE 2770); RNA splicing fa... 54.7 7e-08
dre:323855 c20orf14, fc12b02, prpf6, wu:fa05f07, wu:fc12b02, z... 53.5 2e-07
tpv:TP01_0797 hypothetical protein; K12855 pre-mRNA-processing... 49.3 3e-06
pfa:PF11_0108 U5 snRNP-associated protein, putative; K12855 pr... 48.9 4e-06
xla:447198 prpf6, MGC80263; PRP6 pre-mRNA processing factor 6 ... 47.8 1e-05
mmu:68879 Prpf6, 1190003A07Rik, 2610031L17Rik, ANT-1, MGC11655... 46.6 2e-05
hsa:24148 PRPF6, ANT-1, C20orf14, Prp6, TOM, U5-102K, hPrp6; P... 46.6 2e-05
bbo:BBOV_IV009720 23.m06014; u5 snRNP-associated subunit, puta... 38.1 0.007
cel:Y59A8B.6 hypothetical protein; K12855 pre-mRNA-processing ... 34.3 0.10
hsa:653889 pre-mRNA-processing factor 6-like 28.1 7.8
> tgo:TGME49_005220 U5 snRNP-associated 102 kDa protein, putative
; K12855 pre-mRNA-processing factor 6
Length=985
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 34/51 (66%), Positives = 40/51 (78%), Gaps = 0/51 (0%)
Query 1 FFKTAQAIIRAAIKEGVENINAKRVWSEDAEECLSRGSITVARALYTHALE 51
F T QAI+RA +K GVE +NAKR+W EDAEE LSRGS+ ARALYT A+E
Sbjct 584 FMATCQAIVRATMKVGVEGMNAKRIWKEDAEEALSRGSVATARALYTCAIE 634
> ath:AT4G03430 EMB2770 (EMBRYO DEFECTIVE 2770); RNA splicing
factor, transesterification mechanism; K12855 pre-mRNA-processing
factor 6
Length=1029
Score = 54.7 bits (130), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 27/47 (57%), Positives = 31/47 (65%), Gaps = 0/47 (0%)
Query 4 TAQAIIRAAIKEGVENINAKRVWSEDAEECLSRGSITVARALYTHAL 50
T QAII+ I GVE + KR W DA+EC RGSI ARA+Y HAL
Sbjct 606 TCQAIIKNTIGIGVEEEDRKRTWVADADECKKRGSIETARAIYAHAL 652
> dre:323855 c20orf14, fc12b02, prpf6, wu:fa05f07, wu:fc12b02,
zgc:65913; c20orf14 homolog (H. sapiens); K12855 pre-mRNA-processing
factor 6
Length=944
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 23/48 (47%), Positives = 33/48 (68%), Gaps = 0/48 (0%)
Query 4 TAQAIIRAAIKEGVENINAKRVWSEDAEECLSRGSITVARALYTHALE 51
T Q++IRA I G+E + K W EDA+ C+S G++ ARA+Y HAL+
Sbjct 524 TCQSVIRAVIGIGIEEEDCKHTWMEDADSCVSHGALECARAIYAHALQ 571
> tpv:TP01_0797 hypothetical protein; K12855 pre-mRNA-processing
factor 6
Length=1032
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 26/51 (50%), Positives = 32/51 (62%), Gaps = 0/51 (0%)
Query 1 FFKTAQAIIRAAIKEGVENINAKRVWSEDAEECLSRGSITVARALYTHALE 51
F KTAQ+II+ + GV+ N K VW ED E + GS ARALY +ALE
Sbjct 523 FVKTAQSIIKNTMTIGVDEHNRKSVWLEDGETFVEHGSYECARALYKNALE 573
> pfa:PF11_0108 U5 snRNP-associated protein, putative; K12855
pre-mRNA-processing factor 6
Length=1329
Score = 48.9 bits (115), Expect = 4e-06, Method: Composition-based stats.
Identities = 22/51 (43%), Positives = 32/51 (62%), Gaps = 0/51 (0%)
Query 1 FFKTAQAIIRAAIKEGVENINAKRVWSEDAEECLSRGSITVARALYTHALE 51
F T ++IIR + GVE +N KR++ +DA+ C+ SI AR LY AL+
Sbjct 634 FTHTCESIIRNTMHIGVETLNKKRIYKQDAQNCIHNKSIHTARTLYNEALK 684
> xla:447198 prpf6, MGC80263; PRP6 pre-mRNA processing factor
6 homolog; K12855 pre-mRNA-processing factor 6
Length=948
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 21/48 (43%), Positives = 31/48 (64%), Gaps = 0/48 (0%)
Query 4 TAQAIIRAAIKEGVENINAKRVWSEDAEECLSRGSITVARALYTHALE 51
T QAIIR I G+E + K W EDA+ C++ ++ ARA+Y H+L+
Sbjct 528 TCQAIIRDVIGIGIEEEDRKHTWMEDADSCVAHSALECARAIYAHSLQ 575
> mmu:68879 Prpf6, 1190003A07Rik, 2610031L17Rik, ANT-1, MGC11655,
MGC36967, MGC38351, U5-102K; PRP6 pre-mRNA splicing factor
6 homolog (yeast); K12855 pre-mRNA-processing factor 6
Length=941
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 32/48 (66%), Gaps = 0/48 (0%)
Query 4 TAQAIIRAAIKEGVENINAKRVWSEDAEECLSRGSITVARALYTHALE 51
T QA++RA I G+E + K W EDA+ C++ ++ ARA+Y +AL+
Sbjct 521 TCQAVMRAVIGIGIEEEDRKHTWMEDADSCVAHNALECARAIYAYALQ 568
> hsa:24148 PRPF6, ANT-1, C20orf14, Prp6, TOM, U5-102K, hPrp6;
PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae);
K12855 pre-mRNA-processing factor 6
Length=941
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 32/48 (66%), Gaps = 0/48 (0%)
Query 4 TAQAIIRAAIKEGVENINAKRVWSEDAEECLSRGSITVARALYTHALE 51
T QA++RA I G+E + K W EDA+ C++ ++ ARA+Y +AL+
Sbjct 521 TCQAVMRAVIGIGIEEEDRKHTWMEDADSCVAHNALECARAIYAYALQ 568
> bbo:BBOV_IV009720 23.m06014; u5 snRNP-associated subunit, putaitve;
K12855 pre-mRNA-processing factor 6
Length=1040
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 1 FFKTAQAIIRAAIKEGVENINAKRVWSEDAEECLSRGSITVARALYTHALE 51
F +TAQAII+ + G++ K W ED E + ARA+Y ALE
Sbjct 527 FVQTAQAIIKCTMNIGLDPALLKETWLEDGERMEEKKLFACARAIYRSALE 577
> cel:Y59A8B.6 hypothetical protein; K12855 pre-mRNA-processing
factor 6
Length=968
Score = 34.3 bits (77), Expect = 0.10, Method: Composition-based stats.
Identities = 19/48 (39%), Positives = 25/48 (52%), Gaps = 0/48 (0%)
Query 4 TAQAIIRAAIKEGVENINAKRVWSEDAEECLSRGSITVARALYTHALE 51
T QAIIR I GVE+ + + W DAE + T R +Y AL+
Sbjct 547 TCQAIIRNVIGLGVEDEDKRTTWLADAENFEKEEAFTCVRTVYAIALK 594
> hsa:653889 pre-mRNA-processing factor 6-like
Length=406
Score = 28.1 bits (61), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 10/24 (41%), Positives = 18/24 (75%), Gaps = 0/24 (0%)
Query 28 EDAEECLSRGSITVARALYTHALE 51
EDA+ C++ ++ ARA+Y +AL+
Sbjct 10 EDADSCVAHNALECARAIYAYALQ 33
Lambda K H
0.319 0.128 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2026933688
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40