bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6798_orf2
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
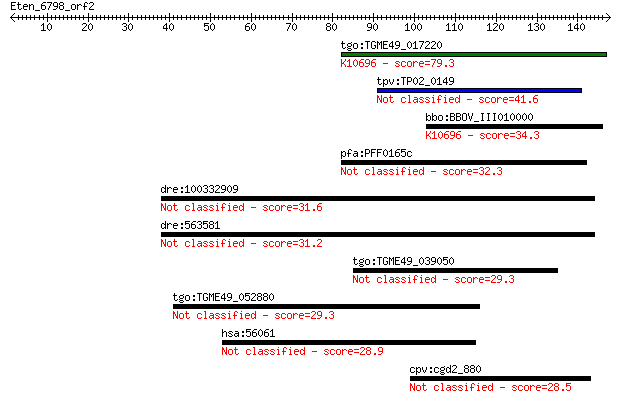
tgo:TGME49_017220 zinc finger (C3HC4 RING finger) protein, put... 79.3 4e-15
tpv:TP02_0149 hypothetical protein 41.6 8e-04
bbo:BBOV_III010000 17.m07868; hypothetical protein; K10696 E3 ... 34.3 0.14
pfa:PFF0165c conserved Plasmodium protein, unknown function 32.3 0.48
dre:100332909 transmembrane and coiled-coil domain family 1-like 31.6 0.94
dre:563581 zgc:153972 31.2 1.3
tgo:TGME49_039050 hypothetical protein 29.3 4.1
tgo:TGME49_052880 hypothetical protein 29.3 5.2
hsa:56061 UBFD1, FLJ38870, FLJ42145, UBPH; ubiquitin family do... 28.9 5.5
cpv:cgd2_880 hypothetical protein 28.5 8.3
> tgo:TGME49_017220 zinc finger (C3HC4 RING finger) protein, putative
; K10696 E3 ubiquitin-protein ligase BRE1 [EC:6.3.2.19]
Length=1027
Score = 79.3 bits (194), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 38/65 (58%), Positives = 53/65 (81%), Gaps = 0/65 (0%)
Query 82 LRIEMQAMQRRLNEQQALEAEVAELRQQYSHVTEELEEVSKAFEERQTHCENLLRQLRER 141
+++E A+QRRL + L E+A LR+QY+ V+EE+EEVSKAFEERQT CE+LL+Q++E+
Sbjct 733 MKLEKDALQRRLADYDQLVVEIAGLREQYARVSEEVEEVSKAFEERQTFCEHLLKQVKEK 792
Query 142 DEAAA 146
DEA A
Sbjct 793 DEALA 797
> tpv:TP02_0149 hypothetical protein
Length=654
Score = 41.6 bits (96), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 35/50 (70%), Gaps = 0/50 (0%)
Query 91 RRLNEQQALEAEVAELRQQYSHVTEELEEVSKAFEERQTHCENLLRQLRE 140
+ L+ + + E E+ ++ +++EELE+VS AFE++Q C++L+RQ+ E
Sbjct 419 KNLDSKSSAENELDFYKEHLLNISEELEDVSSAFEKKQRMCDDLMRQINE 468
> bbo:BBOV_III010000 17.m07868; hypothetical protein; K10696 E3
ubiquitin-protein ligase BRE1 [EC:6.3.2.19]
Length=655
Score = 34.3 bits (77), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 21/47 (44%), Positives = 32/47 (68%), Gaps = 4/47 (8%)
Query 103 VAELRQQYSHVTEELEEVSKAFEERQTHCENLLRQLRE----RDEAA 145
VA+ ++Q +++ ELEE+S A+EE+ T E L RQL+E RD+ A
Sbjct 428 VADYQEQLQNLSVELEEISLAYEEKVTENERLQRQLKELNMYRDKCA 474
> pfa:PFF0165c conserved Plasmodium protein, unknown function
Length=1103
Score = 32.3 bits (72), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 41/60 (68%), Gaps = 0/60 (0%)
Query 82 LRIEMQAMQRRLNEQQALEAEVAELRQQYSHVTEELEEVSKAFEERQTHCENLLRQLRER 141
L IE Q ++ LN+++ +E E+ LR+ Y+ + EE+EE++K FE++Q + ++ Q++ +
Sbjct 832 LFIENQKLKEELNKKRNVEEELHSLRKNYNIINEEIEEITKEFEKKQEQVDEMILQIKNK 891
> dre:100332909 transmembrane and coiled-coil domain family 1-like
Length=425
Score = 31.6 bits (70), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 26/106 (24%), Positives = 45/106 (42%), Gaps = 6/106 (5%)
Query 38 AAEKAAAAAASGDSPPAAAAAAPAAAATAAAKDDAGDNGSGESLLRIEMQAMQRRLNEQQ 97
+E ++A SG + + AP ++ GSG L E+Q ++ + Q
Sbjct 282 GSEDDCSSATSGSAGANSTTGAPGGPPSSKGNTLEHGQGSGFDTLLHEIQELK---DNQS 338
Query 98 ALEAEVAELRQQYSHVTEELEEVSKAFEERQTHCENLLRQLRERDE 143
LE L+ SH + ++ +A +E + CE L QL + E
Sbjct 339 RLEESFDNLK---SHYQRDYTDIMQALQEERFRCERLEEQLNDLTE 381
> dre:563581 zgc:153972
Length=656
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 26/106 (24%), Positives = 45/106 (42%), Gaps = 6/106 (5%)
Query 38 AAEKAAAAAASGDSPPAAAAAAPAAAATAAAKDDAGDNGSGESLLRIEMQAMQRRLNEQQ 97
+E ++A SG + + AP ++ GSG L E+Q ++ + Q
Sbjct 421 GSEDDCSSATSGSAGANSTTGAPGGPPSSKGNTLEHGQGSGFDTLLHEIQELK---DNQS 477
Query 98 ALEAEVAELRQQYSHVTEELEEVSKAFEERQTHCENLLRQLRERDE 143
LE L+ SH + ++ +A +E + CE L QL + E
Sbjct 478 RLEESFDNLK---SHYQRDYTDIMQALQEERFRCERLEEQLNDLTE 520
> tgo:TGME49_039050 hypothetical protein
Length=1182
Score = 29.3 bits (64), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 26/50 (52%), Gaps = 3/50 (6%)
Query 85 EMQAMQRRLNEQQALEAEVAELRQQYSHVTEELEEVSKAFEERQTHCENL 134
EMQ MQRR +E LE ++ R + V +EL+E+ K E C L
Sbjct 1049 EMQCMQRRRDE---LEEQLRLQRMAHEQVVKELQELQKRHEHTMALCSQL 1095
> tgo:TGME49_052880 hypothetical protein
Length=3039
Score = 29.3 bits (64), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 28/78 (35%), Positives = 39/78 (50%), Gaps = 4/78 (5%)
Query 41 KAAAAAASGDSPPAAAAAAPA---AAATAAAKDDAGDNGSGESLLRIEMQAMQRRLNEQQ 97
K A A S SPPA A AP A T A G +GE L +E++ + R E+
Sbjct 1561 KTAEAPKSKASPPAKAKLAPVPKNAGGTDARVGYTGVGDTGEKLRELEVEVKELR-AEKA 1619
Query 98 ALEAEVAELRQQYSHVTE 115
AL E A+L+++ S V +
Sbjct 1620 ALLEENAKLKREGSQVVQ 1637
> hsa:56061 UBFD1, FLJ38870, FLJ42145, UBPH; ubiquitin family
domain containing 1
Length=309
Score = 28.9 bits (63), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 17/62 (27%), Positives = 35/62 (56%), Gaps = 1/62 (1%)
Query 53 PAAAAAAPAAAATAAAKDDAGDNGSGESLLRIEMQAMQRRLNEQQALEAEVAELRQQYSH 112
PA PAA A+ + +DAG G+G L+ +++ + + + + L++ +EL+Q+
Sbjct 58 PAQPPGDPAAQASVSNGEDAG-GGAGRELVDLKIIWNKTKHDVKFPLDSTGSELKQKIHS 116
Query 113 VT 114
+T
Sbjct 117 IT 118
> cpv:cgd2_880 hypothetical protein
Length=633
Score = 28.5 bits (62), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 28/44 (63%), Gaps = 0/44 (0%)
Query 99 LEAEVAELRQQYSHVTEELEEVSKAFEERQTHCENLLRQLRERD 142
L+ E E + ++ E EE+S AFEE+ C++L ++L+E++
Sbjct 339 LKLEKGEKEEVIENLMNEYEELSNAFEEKSLECDDLRQKLKEKE 382
Lambda K H
0.305 0.115 0.292
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2938175820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40