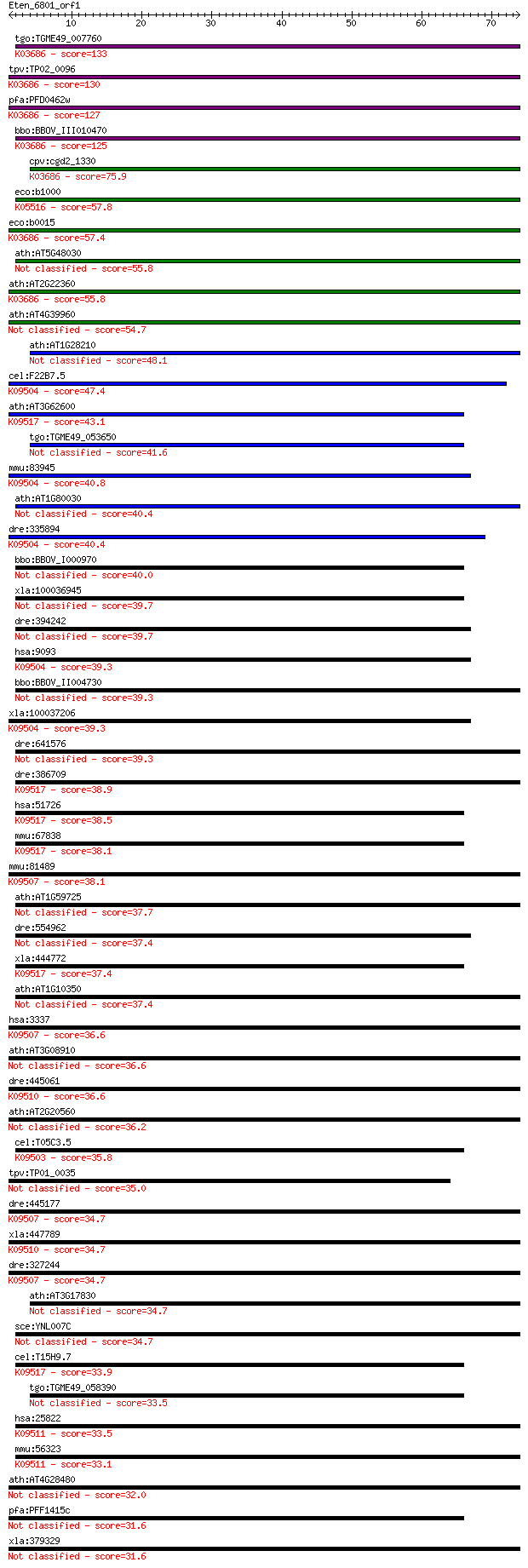
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6801_orf1
Length=73
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_007760 DnaJ domain-containing protein ; K03686 mole... 133 1e-31
tpv:TP02_0096 chaperone protein DnaJ; K03686 molecular chapero... 130 1e-30
pfa:PFD0462w pfj1; DNAJ protein; K03686 molecular chaperone DnaJ 127 1e-29
bbo:BBOV_III010470 17.m07904; dnaJ C terminal region domain co... 125 4e-29
cpv:cgd2_1330 DNAJ protein ; K03686 molecular chaperone DnaJ 75.9 3e-14
eco:b1000 cbpA, ECK0991, JW0985; curved DNA-binding protein, D... 57.8 9e-09
eco:b0015 dnaJ, ECK0015, faa, groP, grpC, JW0014; chaperone HS... 57.4 1e-08
ath:AT5G48030 GFA2; GFA2 (GAMETOPHYTIC FACTOR 2); heat shock p... 55.8 3e-08
ath:AT2G22360 DNAJ heat shock family protein; K03686 molecular... 55.8 3e-08
ath:AT4G39960 DNAJ heat shock family protein 54.7 7e-08
ath:AT1G28210 ATJ1; ATJ1; heat shock protein binding / unfolde... 48.1 6e-06
cel:F22B7.5 dnj-10; DNaJ domain (prokaryotic heat shock protei... 47.4 1e-05
ath:AT3G62600 ATERDJ3B; heat shock protein binding / unfolded ... 43.1 2e-04
tgo:TGME49_053650 DnaJ domain-containing protein 41.6 6e-04
mmu:83945 Dnaja3, 1200003J13Rik, 1810053A11Rik, C81173, Tid-1,... 40.8 0.001
ath:AT1G80030 DNAJ heat shock protein, putative 40.4 0.001
dre:335894 dnaja3a, wu:fj36c02; DnaJ (Hsp40) homolog, subfamil... 40.4 0.001
bbo:BBOV_I000970 16.m00705; DnaJ domain containing protein 40.0 0.002
xla:100036945 hypothetical protein LOC100036945 39.7 0.002
dre:394242 dnaja3b; DnaJ (Hsp40) homolog, subfamily A, member 3B 39.7 0.002
hsa:9093 DNAJA3, FLJ45758, TID1, hTid-1; DnaJ (Hsp40) homolog,... 39.3 0.003
bbo:BBOV_II004730 18.m06396; membrane protein 39.3 0.003
xla:100037206 dnaja3, tid1; DnaJ (Hsp40) homolog, subfamily A,... 39.3 0.003
dre:641576 MGC122979, MGC152710, zgc:152710; zgc:122979 39.3 0.003
dre:386709 dnajb11, cb954; DnaJ (Hsp40) homolog, subfamily B, ... 38.9 0.004
hsa:51726 DNAJB11, ABBP-2, ABBP2, DJ9, EDJ, ERdj3, ERj3, HEDJ,... 38.5 0.006
mmu:67838 Dnajb11, 1810031F23Rik, ABBP-2, AL024055, Dj9, ERdj3... 38.1 0.007
mmu:81489 Dnajb1, 0610007I11Rik, HSPF1, Hsp40; DnaJ (Hsp40) ho... 38.1 0.008
ath:AT1G59725 DNAJ heat shock protein, putative 37.7 0.008
dre:554962 DnaJ (Hsp40) homolog, subfamily A, member 3B 37.4 0.010
xla:444772 dnajb11, MGC81924; DnaJ (Hsp40) homolog, subfamily ... 37.4 0.012
ath:AT1G10350 DNAJ heat shock protein, putative 37.4 0.012
hsa:3337 DNAJB1, HSPF1, Hdj1, Hsp40, RSPH16B, Sis1; DnaJ (Hsp4... 36.6 0.019
ath:AT3G08910 DNAJ heat shock protein, putative 36.6 0.021
dre:445061 dnajb4, zgc:91922; DnaJ (Hsp40) homolog, subfamily ... 36.6 0.022
ath:AT2G20560 DNAJ heat shock family protein 36.2 0.029
cel:T05C3.5 dnj-19; DNaJ domain (prokaryotic heat shock protei... 35.8 0.037
tpv:TP01_0035 chaperone protein DnaJ 35.0 0.064
dre:445177 dnajb1a, dnajb1, hspf1, zf-Hsp40, zgc:101068; DnaJ ... 34.7 0.071
xla:447789 dnajb4, MGC83507; DnaJ (Hsp40) homolog, subfamily B... 34.7 0.073
dre:327244 dnajb1b, dnj-13C, fd19c10, wu:fd19c10, zgc:55492; D... 34.7 0.073
ath:AT3G17830 DNAJ heat shock family protein 34.7 0.079
sce:YNL007C SIS1; Sis1p 34.7 0.080
cel:T15H9.7 dnj-20; DNaJ domain (prokaryotic heat shock protei... 33.9 0.11
tgo:TGME49_058390 DnaJ protein, putative 33.5 0.15
hsa:25822 DNAJB5, Hsc40, KIAA1045; DnaJ (Hsp40) homolog, subfa... 33.5 0.19
mmu:56323 Dnajb5, 1110058L06Rik, AI462558, Hsc40, Hsp40-3; Dna... 33.1 0.20
ath:AT4G28480 DNAJ heat shock family protein 32.0 0.51
pfa:PFF1415c DNAJ domain protein, putative 31.6 0.62
xla:379329 dnaja4.2, MGC52928, dnaja4; DnaJ (Hsp40) homolog, s... 31.6 0.68
> tgo:TGME49_007760 DnaJ domain-containing protein ; K03686 molecular
chaperone DnaJ
Length=1519
Score = 133 bits (335), Expect = 1e-31, Method: Composition-based stats.
Identities = 57/72 (79%), Positives = 65/72 (90%), Gaps = 0/72 (0%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQ 61
ID+PRGVR GMQMRVPNQGH GLRGGKPGHLFVSIN++PH +FRW+DDD+H DVPLS+KQ
Sbjct 1213 IDIPRGVRQGMQMRVPNQGHMGLRGGKPGHLFVSINVKPHDIFRWVDDDIHIDVPLSIKQ 1272
Query 62 CLLGGRVAVPTL 73
CLLGG V + TL
Sbjct 1273 CLLGGSVEISTL 1284
> tpv:TP02_0096 chaperone protein DnaJ; K03686 molecular chaperone
DnaJ
Length=458
Score = 130 bits (326), Expect = 1e-30, Method: Composition-based stats.
Identities = 51/73 (69%), Positives = 66/73 (90%), Gaps = 0/73 (0%)
Query 1 TIDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLK 60
++D+P GVR+GMQMR+PNQGH G+RGGK GHLFV+INI+PH +F+WIDDD+H VP+SLK
Sbjct 295 SVDLPPGVRNGMQMRIPNQGHVGVRGGKSGHLFVNINIQPHHIFKWIDDDIHVHVPISLK 354
Query 61 QCLLGGRVAVPTL 73
QCLLGG +++PTL
Sbjct 355 QCLLGGNISIPTL 367
> pfa:PFD0462w pfj1; DNAJ protein; K03686 molecular chaperone
DnaJ
Length=672
Score = 127 bits (318), Expect = 1e-29, Method: Composition-based stats.
Identities = 49/73 (67%), Positives = 63/73 (86%), Gaps = 0/73 (0%)
Query 1 TIDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLK 60
T+D+P G++ GMQMR+PNQGH G RGGK GHLFV+INI PH +F+W+DD+++ DVPL++K
Sbjct 273 TLDIPPGIKKGMQMRIPNQGHCGYRGGKSGHLFVTINIEPHKIFKWVDDNIYVDVPLTIK 332
Query 61 QCLLGGRVAVPTL 73
QCLLGG V VPTL
Sbjct 333 QCLLGGLVTVPTL 345
> bbo:BBOV_III010470 17.m07904; dnaJ C terminal region domain
containing protein; K03686 molecular chaperone DnaJ
Length=395
Score = 125 bits (313), Expect = 4e-29, Method: Composition-based stats.
Identities = 53/72 (73%), Positives = 63/72 (87%), Gaps = 0/72 (0%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQ 61
I++P GVR GMQMRVPNQGH G+RGGK GHLFV+INI+PH +F+WIDDD+H +P+SLKQ
Sbjct 229 INIPAGVRQGMQMRVPNQGHVGVRGGKSGHLFVNINIQPHQIFKWIDDDIHVHIPISLKQ 288
Query 62 CLLGGRVAVPTL 73
CLLGG V VPTL
Sbjct 289 CLLGGNVVVPTL 300
> cpv:cgd2_1330 DNAJ protein ; K03686 molecular chaperone DnaJ
Length=457
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 27/70 (38%), Positives = 48/70 (68%), Gaps = 2/70 (2%)
Query 4 VPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQCL 63
+PRG + G Q+++ ++G+ G G LF+ +NI+P+ F+WI D++H D+P+S+ C+
Sbjct 307 IPRGTKDGTQLKLSSEGN--FVSGNYGDLFIKVNIKPNTKFKWIKDNIHVDIPISINTCI 364
Query 64 LGGRVAVPTL 73
GG + VP+L
Sbjct 365 FGGEIVVPSL 374
> eco:b1000 cbpA, ECK0991, JW0985; curved DNA-binding protein,
DnaJ homologue that functions as a co-chaperone of DnaK; K05516
curved DNA-binding protein
Length=306
Score = 57.8 bits (138), Expect = 9e-09, Method: Composition-based stats.
Identities = 29/72 (40%), Positives = 43/72 (59%), Gaps = 0/72 (0%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQ 61
+ +P GV +G ++R+ QG G GG G L++ I+I PH +F + DL VP+S +
Sbjct 162 VKIPAGVGNGQRIRLKGQGTPGENGGPNGDLWLVIHIAPHPLFDIVGQDLEIVVPVSPWE 221
Query 62 CLLGGRVAVPTL 73
LG +V VPTL
Sbjct 222 AALGAKVTVPTL 233
> eco:b0015 dnaJ, ECK0015, faa, groP, grpC, JW0014; chaperone
HSP40, co-chaperone with DnaK; K03686 molecular chaperone DnaJ
Length=376
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 24/73 (32%), Positives = 43/73 (58%), Gaps = 0/73 (0%)
Query 1 TIDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLK 60
++ +P GV G ++R+ +G AG G G L+V + ++ H +F ++L+ +VP++
Sbjct 213 SVKIPAGVDTGDRIRLAGEGEAGEHGAPAGDLYVQVQVKQHPIFEREGNNLYCEVPINFA 272
Query 61 QCLLGGRVAVPTL 73
LGG + VPTL
Sbjct 273 MAALGGEIEVPTL 285
> ath:AT5G48030 GFA2; GFA2 (GAMETOPHYTIC FACTOR 2); heat shock
protein binding / unfolded protein binding
Length=456
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 27/72 (37%), Positives = 41/72 (56%), Gaps = 0/72 (0%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQ 61
+ + GV + ++V G A G +PG L+V++ +R VFR D+H D LS+ Q
Sbjct 308 VTIDPGVDNSDTLKVARVGGADPEGDQPGDLYVTLKVREDPVFRREGSDIHVDAVLSVTQ 367
Query 62 CLLGGRVAVPTL 73
+LGG + VPTL
Sbjct 368 AILGGTIQVPTL 379
> ath:AT2G22360 DNAJ heat shock family protein; K03686 molecular
chaperone DnaJ
Length=442
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 27/73 (36%), Positives = 42/73 (57%), Gaps = 0/73 (0%)
Query 1 TIDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLK 60
++ VP GV G ++RV +G+AG RGG PG LFV I + P + + D ++ +S
Sbjct 296 SLKVPAGVDSGSRLRVRGEGNAGKRGGSPGDLFVVIEVIPDPILKRDDTNILYTCKISYI 355
Query 61 QCLLGGRVAVPTL 73
+LG + VPT+
Sbjct 356 DAILGTTLKVPTV 368
> ath:AT4G39960 DNAJ heat shock family protein
Length=447
Score = 54.7 bits (130), Expect = 7e-08, Method: Composition-based stats.
Identities = 27/73 (36%), Positives = 41/73 (56%), Gaps = 0/73 (0%)
Query 1 TIDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLK 60
++ VP GV G ++RV +G+AG RGG PG LF I + P V + D ++ +S
Sbjct 302 SLKVPAGVDSGSRLRVRGEGNAGKRGGSPGDLFAVIEVIPDPVLKRDDTNILYTCKISYV 361
Query 61 QCLLGGRVAVPTL 73
+LG + VPT+
Sbjct 362 DAILGTTLKVPTV 374
> ath:AT1G28210 ATJ1; ATJ1; heat shock protein binding / unfolded
protein binding
Length=427
Score = 48.1 bits (113), Expect = 6e-06, Method: Composition-based stats.
Identities = 21/70 (30%), Positives = 37/70 (52%), Gaps = 0/70 (0%)
Query 4 VPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQCL 63
+P GV + + G+ R +PG+L++ + + + F D++ D +S Q +
Sbjct 254 IPGGVESEATITIVGAGNVSSRTSQPGNLYIKLKVANDSTFTRDGSDIYVDANISFTQAI 313
Query 64 LGGRVAVPTL 73
LGG+V VPTL
Sbjct 314 LGGKVVVPTL 323
> cel:F22B7.5 dnj-10; DNaJ domain (prokaryotic heat shock protein)
family member (dnj-10); K09504 DnaJ homolog subfamily A
member 3
Length=446
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 27/71 (38%), Positives = 37/71 (52%), Gaps = 9/71 (12%)
Query 1 TIDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLK 60
+ +VP G +G ++ + GK LFV N+ P FR DD+H DV +SL
Sbjct 261 SFNVPAGTNNGDSLK--------FQVGK-NQLFVRFNVAPSLKFRREKDDIHCDVDISLA 311
Query 61 QCLLGGRVAVP 71
Q +LGG V VP
Sbjct 312 QAVLGGTVKVP 322
> ath:AT3G62600 ATERDJ3B; heat shock protein binding / unfolded
protein binding; K09517 DnaJ homolog subfamily B member 11
Length=346
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 23/65 (35%), Positives = 36/65 (55%), Gaps = 1/65 (1%)
Query 1 TIDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLK 60
T+D+ +G++ G ++ G L G PG L I PHA FR +DLH +V ++L
Sbjct 213 TVDIEKGMKDGEEVSFYEDGEPIL-DGDPGDLKFRIRTAPHARFRRDGNDLHMNVNITLV 271
Query 61 QCLLG 65
+ L+G
Sbjct 272 EALVG 276
> tgo:TGME49_053650 DnaJ domain-containing protein
Length=403
Score = 41.6 bits (96), Expect = 6e-04, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 32/62 (51%), Gaps = 0/62 (0%)
Query 4 VPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQCL 63
+P GV GM++R+ QG G+ GG G L + + + FR ++L V +S +
Sbjct 257 IPAGVSDGMRLRIKGQGDVGVNGGPSGDLLLKVEVENDTAFRREGNNLLGIVRISYIDAI 316
Query 64 LG 65
LG
Sbjct 317 LG 318
> mmu:83945 Dnaja3, 1200003J13Rik, 1810053A11Rik, C81173, Tid-1,
Tid1l; DnaJ (Hsp40) homolog, subfamily A, member 3; K09504
DnaJ homolog subfamily A member 3
Length=453
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 24/66 (36%), Positives = 36/66 (54%), Gaps = 9/66 (13%)
Query 1 TIDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLK 60
TI VP GV G +R+P G R +FV+ ++ VFR D+H+D+ +S+
Sbjct 305 TIPVPAGVEDGQTVRMP----VGKR-----EIFVTFRVQKSPVFRRDGADIHSDLFISIA 355
Query 61 QCLLGG 66
Q +LGG
Sbjct 356 QAILGG 361
> ath:AT1G80030 DNAJ heat shock protein, putative
Length=500
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 25/75 (33%), Positives = 40/75 (53%), Gaps = 6/75 (8%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDD---LHADVPLS 58
+ +P GV G +RV +G +G RGG PG L+V +++ R I+ D L + + +S
Sbjct 296 VKIPPGVSAGSILRVAGEGDSGPRGGPPGDLYVYLDVED---VRGIERDGINLLSTLSIS 352
Query 59 LKQCLLGGRVAVPTL 73
+LG V V T+
Sbjct 353 YLDAILGAVVKVKTV 367
> dre:335894 dnaja3a, wu:fj36c02; DnaJ (Hsp40) homolog, subfamily
A, member 3A; K09504 DnaJ homolog subfamily A member 3
Length=453
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 21/68 (30%), Positives = 37/68 (54%), Gaps = 9/68 (13%)
Query 1 TIDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLK 60
T+ VP G+ G +R+P GK +F++ ++ +FR D+H+DV +S+
Sbjct 305 TVPVPAGIEDGQTVRMPV--------GKK-EIFITFKVQKSPIFRRDGADIHSDVMISVA 355
Query 61 QCLLGGRV 68
Q +LGG +
Sbjct 356 QAILGGTI 363
> bbo:BBOV_I000970 16.m00705; DnaJ domain containing protein
Length=371
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 36/64 (56%), Gaps = 1/64 (1%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQ 61
I + G +HG Q+ + +G G GH+ +I +PH ++R +DL+ + ++LK+
Sbjct 241 ITIEPGAKHGQQIIIEGRGQE-RPGLHRGHIIFTIEQKPHHIYRREGNDLYRTLDITLKE 299
Query 62 CLLG 65
LLG
Sbjct 300 ALLG 303
> xla:100036945 hypothetical protein LOC100036945
Length=358
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 21/64 (32%), Positives = 35/64 (54%), Gaps = 1/64 (1%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQ 61
+++ GVR GM+ +G + G+PG L I + H +F DDL+ +V +SL +
Sbjct 210 VEIEPGVRDGMEYPFIGEGEPHI-DGEPGDLRFRIKVLKHPIFERRGDDLYTNVSISLVE 268
Query 62 CLLG 65
L+G
Sbjct 269 ALIG 272
> dre:394242 dnaja3b; DnaJ (Hsp40) homolog, subfamily A, member
3B
Length=474
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 36/65 (55%), Gaps = 9/65 (13%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQ 61
+ VP G+ G ++VP GK H++++ ++ VFR D+H+DV +S+ Q
Sbjct 299 VPVPAGIADGQTVKVPV--------GKK-HMYITFRVQKSPVFRRDGADIHSDVLISIAQ 349
Query 62 CLLGG 66
+LGG
Sbjct 350 AILGG 354
> hsa:9093 DNAJA3, FLJ45758, TID1, hTid-1; DnaJ (Hsp40) homolog,
subfamily A, member 3; K09504 DnaJ homolog subfamily A member
3
Length=453
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 23/65 (35%), Positives = 35/65 (53%), Gaps = 9/65 (13%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQ 61
I VP GV G +R+P G R +F++ ++ VFR D+H+D+ +S+ Q
Sbjct 306 IPVPAGVEDGQTVRMP----VGKR-----EIFITFRVQKSPVFRRDGADIHSDLFISIAQ 356
Query 62 CLLGG 66
LLGG
Sbjct 357 ALLGG 361
> bbo:BBOV_II004730 18.m06396; membrane protein
Length=510
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 22/73 (30%), Positives = 38/73 (52%), Gaps = 2/73 (2%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWID-DDLHADVPLSLK 60
I +P G++ G ++V +GH+ +PG+L+V I+ P+ +ID D ++ L
Sbjct 381 IKIPAGIKVGSTLKVKGKGHSSGYSSRPGNLYVKISSSPNE-HEYIDMDKVYTTATLDYV 439
Query 61 QCLLGGRVAVPTL 73
+LG V V T
Sbjct 440 SAILGDTVNVYTF 452
> xla:100037206 dnaja3, tid1; DnaJ (Hsp40) homolog, subfamily
A, member 3; K09504 DnaJ homolog subfamily A member 3
Length=457
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 22/66 (33%), Positives = 37/66 (56%), Gaps = 9/66 (13%)
Query 1 TIDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLK 60
T+ VP GV +G +R+P GK +F++ ++ VFR D+H+D+ +S+
Sbjct 280 TVPVPAGVENGQTVRMPV--------GKK-EIFITFRVQKSPVFRRDGADIHSDLFISVA 330
Query 61 QCLLGG 66
Q +LGG
Sbjct 331 QAVLGG 336
> dre:641576 MGC122979, MGC152710, zgc:152710; zgc:122979
Length=360
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 24/72 (33%), Positives = 37/72 (51%), Gaps = 1/72 (1%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQ 61
++V +G + G ++ PN+GH L G P L I + HA FR + ++L++
Sbjct 225 VEVKKGWKEGTRITFPNEGHQML-GHAPNDLAFVIKEKKHAHFRRDGSHIVYTCTITLRE 283
Query 62 CLLGGRVAVPTL 73
L G V VPTL
Sbjct 284 ALCGCTVNVPTL 295
> dre:386709 dnajb11, cb954; DnaJ (Hsp40) homolog, subfamily B,
member 11; K09517 DnaJ homolog subfamily B member 11
Length=360
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 23/72 (31%), Positives = 38/72 (52%), Gaps = 1/72 (1%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQ 61
+++ +GVR M+ +G + G+PG L I + H VF DDL+ +V +SL +
Sbjct 212 VEIEQGVRDEMEYPFIGEGEPHI-DGEPGDLRFRIKVLKHPVFERRGDDLYTNVTISLVE 270
Query 62 CLLGGRVAVPTL 73
L+G V + L
Sbjct 271 ALVGFEVDITHL 282
> hsa:51726 DNAJB11, ABBP-2, ABBP2, DJ9, EDJ, ERdj3, ERj3, HEDJ,
PRO1080, UNQ537, hDj9; DnaJ (Hsp40) homolog, subfamily B,
member 11; K09517 DnaJ homolog subfamily B member 11
Length=358
Score = 38.5 bits (88), Expect = 0.006, Method: Composition-based stats.
Identities = 21/64 (32%), Positives = 35/64 (54%), Gaps = 1/64 (1%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQ 61
+++ GVR GM+ +G + G+PG L I + H +F DDL+ +V +SL +
Sbjct 210 VEIEPGVRDGMEYPFIGEGEPHV-DGEPGDLRFRIKVVKHPIFERRGDDLYTNVTISLVE 268
Query 62 CLLG 65
L+G
Sbjct 269 SLVG 272
> mmu:67838 Dnajb11, 1810031F23Rik, ABBP-2, AL024055, Dj9, ERdj3,
ERj3p; DnaJ (Hsp40) homolog, subfamily B, member 11; K09517
DnaJ homolog subfamily B member 11
Length=358
Score = 38.1 bits (87), Expect = 0.007, Method: Composition-based stats.
Identities = 21/64 (32%), Positives = 35/64 (54%), Gaps = 1/64 (1%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQ 61
+++ GVR GM+ +G + G+PG L I + H +F DDL+ +V +SL +
Sbjct 210 VEIEPGVRDGMEYPFIGEGEPHV-DGEPGDLRFRIKVVKHRIFERRGDDLYTNVTVSLVE 268
Query 62 CLLG 65
L+G
Sbjct 269 ALVG 272
> mmu:81489 Dnajb1, 0610007I11Rik, HSPF1, Hsp40; DnaJ (Hsp40)
homolog, subfamily B, member 1; K09507 DnaJ homolog subfamily
B member 1
Length=340
Score = 38.1 bits (87), Expect = 0.008, Method: Composition-based stats.
Identities = 22/73 (30%), Positives = 36/73 (49%), Gaps = 1/73 (1%)
Query 1 TIDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLK 60
TI+V RG + G ++ P +G P + + +PH +F+ D+ +SL+
Sbjct 205 TIEVKRGWKEGTKITFPKEGDQ-TSNNIPADIVFVLKDKPHNIFKRDGSDVIYPARISLR 263
Query 61 QCLLGGRVAVPTL 73
+ L G V VPTL
Sbjct 264 EALCGCTVNVPTL 276
> ath:AT1G59725 DNAJ heat shock protein, putative
Length=331
Score = 37.7 bits (86), Expect = 0.008, Method: Composition-based stats.
Identities = 22/72 (30%), Positives = 39/72 (54%), Gaps = 1/72 (1%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQ 61
ID+ G + G ++ P +G+ G P L I+ +PH+V++ +DL D +SL +
Sbjct 196 IDITPGWKKGTKITFPEKGNQE-PGVTPADLIFVIDEKPHSVYKRDGNDLIVDKKVSLLE 254
Query 62 CLLGGRVAVPTL 73
L G +++ TL
Sbjct 255 ALTGITLSLTTL 266
> dre:554962 DnaJ (Hsp40) homolog, subfamily A, member 3B
Length=474
Score = 37.4 bits (85), Expect = 0.010, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 36/65 (55%), Gaps = 9/65 (13%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQ 61
+ VP G+ G ++VP GK +++++ ++ VFR D+H+DV +S+ Q
Sbjct 299 VPVPAGIADGQTVKVPV--------GKK-YMYITFRVQKSPVFRRDGADIHSDVMISIAQ 349
Query 62 CLLGG 66
+LGG
Sbjct 350 AILGG 354
> xla:444772 dnajb11, MGC81924; DnaJ (Hsp40) homolog, subfamily
B, member 11; K09517 DnaJ homolog subfamily B member 11
Length=360
Score = 37.4 bits (85), Expect = 0.012, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 34/64 (53%), Gaps = 1/64 (1%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQ 61
+++ GVR M+ +G + G+PG L I + H +F DDL+ +V +SL +
Sbjct 212 VEIEPGVRDSMEYPFIGEGEPHI-DGEPGDLRFRIKVLKHPIFERRGDDLYTNVSISLVE 270
Query 62 CLLG 65
L+G
Sbjct 271 ALIG 274
> ath:AT1G10350 DNAJ heat shock protein, putative
Length=349
Score = 37.4 bits (85), Expect = 0.012, Method: Composition-based stats.
Identities = 22/72 (30%), Positives = 38/72 (52%), Gaps = 1/72 (1%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQ 61
ID+ G + G ++ P +G+ G P L ++ +PH+VF+ +DL + +SL
Sbjct 215 IDIKPGWKKGTKITFPEKGNQE-PGVTPADLIFVVDEKPHSVFKRDGNDLILEKKVSLID 273
Query 62 CLLGGRVAVPTL 73
L G ++V TL
Sbjct 274 ALTGLTISVTTL 285
> hsa:3337 DNAJB1, HSPF1, Hdj1, Hsp40, RSPH16B, Sis1; DnaJ (Hsp40)
homolog, subfamily B, member 1; K09507 DnaJ homolog subfamily
B member 1
Length=340
Score = 36.6 bits (83), Expect = 0.019, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 36/73 (49%), Gaps = 1/73 (1%)
Query 1 TIDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLK 60
TI+V +G + G ++ P +G P + + +PH +F+ D+ +SL+
Sbjct 205 TIEVKKGWKEGTKITFPKEGDQ-TSNNIPADIVFVLKDKPHNIFKRDGSDVIYPARISLR 263
Query 61 QCLLGGRVAVPTL 73
+ L G V VPTL
Sbjct 264 EALCGCTVNVPTL 276
> ath:AT3G08910 DNAJ heat shock protein, putative
Length=323
Score = 36.6 bits (83), Expect = 0.021, Method: Composition-based stats.
Identities = 23/73 (31%), Positives = 37/73 (50%), Gaps = 1/73 (1%)
Query 1 TIDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLK 60
TI++ G + G ++ P +G+ RG P L ++ +PHAVF+ +DL + L
Sbjct 187 TIEIKPGWKKGTKITFPEKGNEQ-RGIIPSDLVFIVDEKPHAVFKRDGNDLVMTQKIPLV 245
Query 61 QCLLGGRVAVPTL 73
+ L G V TL
Sbjct 246 EALTGYTAQVSTL 258
> dre:445061 dnajb4, zgc:91922; DnaJ (Hsp40) homolog, subfamily
B, member 4; K09510 DnaJ homolog subfamily B member 4
Length=340
Score = 36.6 bits (83), Expect = 0.022, Method: Composition-based stats.
Identities = 23/73 (31%), Positives = 35/73 (47%), Gaps = 1/73 (1%)
Query 1 TIDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLK 60
TI++ RG + G ++ P +G P + I +PH FR D+ V +SL+
Sbjct 204 TIEIKRGWKEGTKITFPREGDE-TPNTIPADIVFVIKDKPHGHFRREGSDIVYPVRVSLR 262
Query 61 QCLLGGRVAVPTL 73
Q L G V V T+
Sbjct 263 QSLCGCSVTVSTI 275
> ath:AT2G20560 DNAJ heat shock family protein
Length=337
Score = 36.2 bits (82), Expect = 0.029, Method: Composition-based stats.
Identities = 25/73 (34%), Positives = 36/73 (49%), Gaps = 1/73 (1%)
Query 1 TIDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLK 60
TIDV G + G ++ P +G+ G P L I+ +PH VF +DL +SL
Sbjct 203 TIDVKPGWKKGTKITFPEKGNEQ-PGVIPADLVFIIDEKPHPVFTREGNDLIVTQKISLV 261
Query 61 QCLLGGRVAVPTL 73
+ L G V + TL
Sbjct 262 EALTGYTVNLTTL 274
> cel:T05C3.5 dnj-19; DNaJ domain (prokaryotic heat shock protein)
family member (dnj-19); K09503 DnaJ homolog subfamily A
member 2
Length=439
Score = 35.8 bits (81), Expect = 0.037, Method: Composition-based stats.
Identities = 19/64 (29%), Positives = 31/64 (48%), Gaps = 0/64 (0%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQ 61
+ V G++H ++ G G+PG + + I + H +F+ DDLH LSL +
Sbjct 245 VHVLPGMKHNDKITFKGDGDQSDPDGEPGDVVIVIQQKDHDIFKRDGDDLHMTKKLSLNE 304
Query 62 CLLG 65
L G
Sbjct 305 ALCG 308
> tpv:TP01_0035 chaperone protein DnaJ
Length=383
Score = 35.0 bits (79), Expect = 0.064, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 33/63 (52%), Gaps = 1/63 (1%)
Query 1 TIDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLK 60
T++V G ++ + + +G G K G L + +PH VFR +DLH V +SLK
Sbjct 251 TVNVEPGHKNKQNIVMEGRGQEK-PGMKRGDLVFVVTEKPHDVFRREGNDLHCKVDISLK 309
Query 61 QCL 63
+ L
Sbjct 310 ESL 312
> dre:445177 dnajb1a, dnajb1, hspf1, zf-Hsp40, zgc:101068; DnaJ
(Hsp40) homolog, subfamily B, member 1a; K09507 DnaJ homolog
subfamily B member 1
Length=335
Score = 34.7 bits (78), Expect = 0.071, Method: Composition-based stats.
Identities = 20/73 (27%), Positives = 35/73 (47%), Gaps = 1/73 (1%)
Query 1 TIDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLK 60
T+D+ RG + G ++ P +G P + + + H+VFR D+ +SL+
Sbjct 200 TVDIKRGWKEGTKITFPKEGDE-TPTNIPADIVFVVKDKIHSVFRRDGSDIIYPARISLR 258
Query 61 QCLLGGRVAVPTL 73
+ L G + PTL
Sbjct 259 EALCGCTINAPTL 271
> xla:447789 dnajb4, MGC83507; DnaJ (Hsp40) homolog, subfamily
B, member 4; K09510 DnaJ homolog subfamily B member 4
Length=339
Score = 34.7 bits (78), Expect = 0.073, Method: Composition-based stats.
Identities = 20/76 (26%), Positives = 39/76 (51%), Gaps = 7/76 (9%)
Query 1 TIDVPRGVRHGMQMRVPNQG---HAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPL 57
TI++ +G + G ++ P +G H + P + + +PHA F+ ++ + +
Sbjct 203 TIEIKKGWKEGTKITFPREGDETHMTI----PADIVFVVKDKPHAHFKRDGSNIVSPARI 258
Query 58 SLKQCLLGGRVAVPTL 73
SL++ L G + VPTL
Sbjct 259 SLREALCGCSINVPTL 274
> dre:327244 dnajb1b, dnj-13C, fd19c10, wu:fd19c10, zgc:55492;
DnaJ (Hsp40) homolog, subfamily B, member 1b; K09507 DnaJ homolog
subfamily B member 1
Length=337
Score = 34.7 bits (78), Expect = 0.073, Method: Composition-based stats.
Identities = 19/73 (26%), Positives = 36/73 (49%), Gaps = 1/73 (1%)
Query 1 TIDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLK 60
T++V +G + G ++ P +G P + + +PH V++ D+ ++LK
Sbjct 201 TVEVKKGWKEGTKITFPREGDE-TPSNIPADVVFVLKDKPHPVYKRDGSDIIYPAKITLK 259
Query 61 QCLLGGRVAVPTL 73
+ L G + VPTL
Sbjct 260 EALCGCVINVPTL 272
> ath:AT3G17830 DNAJ heat shock family protein
Length=517
Score = 34.7 bits (78), Expect = 0.079, Method: Composition-based stats.
Identities = 18/70 (25%), Positives = 33/70 (47%), Gaps = 0/70 (0%)
Query 4 VPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQCL 63
VP GV MR+ +G+ R G+ G LF+ + + R +L++++ + +
Sbjct 288 VPPGVSDRATMRIQGEGNMDKRSGRAGDLFIVLQVDEKRGIRREGLNLYSNINIDFTDAI 347
Query 64 LGGRVAVPTL 73
LG V T+
Sbjct 348 LGATTKVETV 357
> sce:YNL007C SIS1; Sis1p
Length=352
Score = 34.7 bits (78), Expect = 0.080, Method: Composition-based stats.
Identities = 22/72 (30%), Positives = 34/72 (47%), Gaps = 0/72 (0%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQ 61
I + G + G ++ NQG + G+ L I + H F+ DDL +PLS K+
Sbjct 219 IQLKPGWKAGTKITYKNQGDYNPQTGRRKTLQFVIQEKSHPNFKRDGDDLIYTLPLSFKE 278
Query 62 CLLGGRVAVPTL 73
LLG + T+
Sbjct 279 SLLGFSKTIQTI 290
> cel:T15H9.7 dnj-20; DNaJ domain (prokaryotic heat shock protein)
family member (dnj-20); K09517 DnaJ homolog subfamily B
member 11
Length=355
Score = 33.9 bits (76), Expect = 0.11, Method: Composition-based stats.
Identities = 21/64 (32%), Positives = 31/64 (48%), Gaps = 1/64 (1%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQ 61
++V G +G Q +G + G PG L I I+ H F DDL+ +V +SL+
Sbjct 209 VEVEVGADNGHQQIFHGEGEPHIEG-DPGDLKFKIRIQKHPRFERKGDDLYTNVTISLQD 267
Query 62 CLLG 65
L G
Sbjct 268 ALNG 271
> tgo:TGME49_058390 DnaJ protein, putative
Length=397
Score = 33.5 bits (75), Expect = 0.15, Method: Composition-based stats.
Identities = 21/62 (33%), Positives = 31/62 (50%), Gaps = 1/62 (1%)
Query 4 VPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQCL 63
V G+R G ++ G L G +PG L + I PH + I DDL + +SL + L
Sbjct 254 VEAGMRDGDEIVFDGVGEQKL-GHEPGDLVLVIQELPHKRYSRIGDDLEMSIRISLLEAL 312
Query 64 LG 65
+G
Sbjct 313 VG 314
> hsa:25822 DNAJB5, Hsc40, KIAA1045; DnaJ (Hsp40) homolog, subfamily
B, member 5; K09511 DnaJ homolog subfamily B member 5
Length=382
Score = 33.5 bits (75), Expect = 0.19, Method: Composition-based stats.
Identities = 21/72 (29%), Positives = 35/72 (48%), Gaps = 1/72 (1%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQ 61
I + RG + G ++ P +G A P + + +PHA FR ++ +SLK+
Sbjct 247 IVIKRGWKEGTKITFPKEGDA-TPDNIPADIVFVLKDKPHAHFRRDGTNVLYSALISLKE 305
Query 62 CLLGGRVAVPTL 73
L G V +PT+
Sbjct 306 ALCGCTVNIPTI 317
> mmu:56323 Dnajb5, 1110058L06Rik, AI462558, Hsc40, Hsp40-3; DnaJ
(Hsp40) homolog, subfamily B, member 5; K09511 DnaJ homolog
subfamily B member 5
Length=348
Score = 33.1 bits (74), Expect = 0.20, Method: Composition-based stats.
Identities = 21/72 (29%), Positives = 35/72 (48%), Gaps = 1/72 (1%)
Query 2 IDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLKQ 61
I + RG + G ++ P +G A P + + +PHA FR ++ +SLK+
Sbjct 213 IVIKRGWKEGTKITFPKEGDA-TPDNIPADIVFVLKDKPHAHFRRDGTNVLYSALISLKE 271
Query 62 CLLGGRVAVPTL 73
L G V +PT+
Sbjct 272 ALCGCTVNIPTI 283
> ath:AT4G28480 DNAJ heat shock family protein
Length=290
Score = 32.0 bits (71), Expect = 0.51, Method: Composition-based stats.
Identities = 24/75 (32%), Positives = 34/75 (45%), Gaps = 5/75 (6%)
Query 1 TIDVPRGVRHGMQMRVPNQG--HAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLS 58
TI V G + G ++ P +G H G+ P L I+ +PH VF +DL +S
Sbjct 156 TIGVKPGWKKGTKITFPEKGNEHPGV---IPADLVFIIDEKPHPVFTREGNDLIVTQKVS 212
Query 59 LKQCLLGGRVAVPTL 73
L L G + TL
Sbjct 213 LADALTGYTANIATL 227
> pfa:PFF1415c DNAJ domain protein, putative
Length=380
Score = 31.6 bits (70), Expect = 0.62, Method: Composition-based stats.
Identities = 14/65 (21%), Positives = 36/65 (55%), Gaps = 1/65 (1%)
Query 1 TIDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLK 60
T+++ +G+++ ++ +G + G + G + + + H ++ +++DLH +SLK
Sbjct 248 TLEIEKGMKNNDKIVFEKKGKQEI-GYENGDIIFIVQTKKHKIYERVNNDLHQIYEISLK 306
Query 61 QCLLG 65
L+G
Sbjct 307 DALIG 311
> xla:379329 dnaja4.2, MGC52928, dnaja4; DnaJ (Hsp40) homolog,
subfamily A, member 4, gene 2
Length=402
Score = 31.6 bits (70), Expect = 0.68, Method: Composition-based stats.
Identities = 19/73 (26%), Positives = 36/73 (49%), Gaps = 1/73 (1%)
Query 1 TIDVPRGVRHGMQMRVPNQGHAGLRGGKPGHLFVSINIRPHAVFRWIDDDLHADVPLSLK 60
T+ + +G++ G ++ +G G +PG + + + + H VF+ DL + + L
Sbjct 214 TVHIDKGMKSGQKIIFHEEGDQA-PGLQPGDIIIVLEQKVHPVFQRKGHDLVMKMEIQLA 272
Query 61 QCLLGGRVAVPTL 73
L G R +V TL
Sbjct 273 DALCGCRQSVKTL 285
Lambda K H
0.326 0.146 0.474
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2011623444
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40