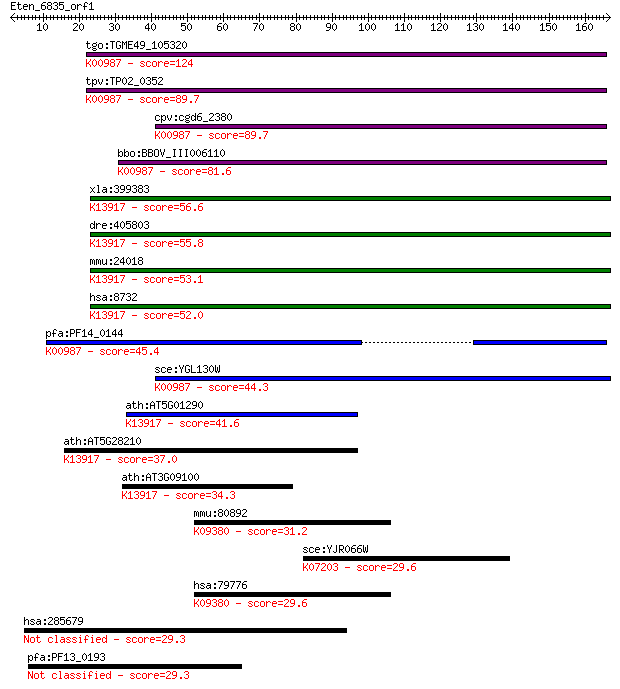
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6835_orf1
Length=166
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_105320 mRNA capping enzyme, putative (EC:2.7.7.50);... 124 2e-28
tpv:TP02_0352 mRNA capping enzyme; K00987 mRNA guanylyltransfe... 89.7 4e-18
cpv:cgd6_2380 mRNA capping enzyme; RNA guanylyltransferase Ceg... 89.7 4e-18
bbo:BBOV_III006110 17.m07541; mRNA capping enzyme, C-terminal ... 81.6 1e-15
xla:399383 rngtt, xcap1b; RNA guanylyltransferase and 5'-phosp... 56.6 4e-08
dre:405803 rngtt, MGC76886, zgc:76886; RNA guanylyltransferase... 55.8 6e-08
mmu:24018 Rngtt, AU020997; RNA guanylyltransferase and 5'-phos... 53.1 4e-07
hsa:8732 RNGTT, CAP1A, DKFZp686J2031, HCE, HCE1, hCAP; RNA gua... 52.0 1e-06
pfa:PF14_0144 pgt1; RNA guanylyltransferase; K00987 mRNA guany... 45.4 1e-04
sce:YGL130W CEG1; Alpha (guanylyltransferase) subunit of the m... 44.3 2e-04
ath:AT5G01290 mRNA guanylyltransferase/ phosphatase/ polynucle... 41.6 0.001
ath:AT5G28210 mRNA capping enzyme family protein; K13917 mRNA-... 37.0 0.028
ath:AT3G09100 mRNA capping enzyme family protein; K13917 mRNA-... 34.3 0.22
mmu:80892 Zfhx4, A930021B15, C130041O22Rik, Zfh-4, Zfh4; zinc ... 31.2 1.6
sce:YJR066W TOR1, DRR1; PIK-related protein kinase and rapamyc... 29.6 4.3
hsa:79776 ZFHX4, FLJ16514, FLJ20980, ZFH4, ZHF4; zinc finger h... 29.6 4.4
hsa:285679 C5orf60, FLJ35723; chromosome 5 open reading frame 60 29.3
pfa:PF13_0193 MSP7-like protein 29.3 7.2
> tgo:TGME49_105320 mRNA capping enzyme, putative (EC:2.7.7.50);
K00987 mRNA guanylyltransferase [EC:2.7.7.50]
Length=509
Score = 124 bits (311), Expect = 2e-28, Method: Composition-based stats.
Identities = 72/155 (46%), Positives = 101/155 (65%), Gaps = 11/155 (7%)
Query 22 QVREISEMYLPRAADSSEAQEETLLDGELVLDQL-EGAAVWRFLVYDCISIDGNETIQKL 80
+VR I + +LPR E Q+ TLLDGELV+D+L G +V R+L+YD I I+ +E+I++L
Sbjct 156 EVRMIPDKFLPRKGRLHEPQQLTLLDGELVMDRLPNGESVARYLIYDAICIERDESIKEL 215
Query 81 NLLKRLHAVRKQVIEPLLLLQLQQQQQQQTAAAA----------AAAVGDSLPLCSKPPM 130
NL+ RL AV ++V+ PL L+ +++ Q + AA A + + K +
Sbjct 216 NLMGRLAAVAERVVAPLRELEEEERMQSERNEAARESHANDGTGEAQLAKTGRTKGKNSL 275
Query 131 EIYLKDFFEIFDLNSIRCLSLRLPHPSDGIIFTPV 165
EIYLKDFFEIFDL I+ ++LRLPH SDGIIFTPV
Sbjct 276 EIYLKDFFEIFDLLHIQRMALRLPHESDGIIFTPV 310
> tpv:TP02_0352 mRNA capping enzyme; K00987 mRNA guanylyltransferase
[EC:2.7.7.50]
Length=419
Score = 89.7 bits (221), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 57/147 (38%), Positives = 86/147 (58%), Gaps = 13/147 (8%)
Query 22 QVREISEMYLPRAADSSEAQEETLLDGELVLDQL--EGAAVWRFLVYDCISIDGNETIQK 79
+V EI +M LP + S++Q+ TLLDGE+V+D+ + + +R+L YD I I +++ K
Sbjct 89 EVHEI-KMKLPVRGNLSQSQQLTLLDGEVVMDENIEDNSVSYRYLCYDGICIQ-RKSLNK 146
Query 80 LNLLKRLHAVRKQVIEPLLLLQL-QQQQQQQTAAAAAAAVGDSLPLCSKPPMEIYLKDFF 138
+NL++RL V VI PL + + + T + D L EIYLKDFF
Sbjct 147 MNLMERLAFVYTHVIVPLRMAGIYSSTPNKPTDGTDNSEAYDKL--------EIYLKDFF 198
Query 139 EIFDLNSIRCLSLRLPHPSDGIIFTPV 165
+I + I +S++LPH SDG+IFTPV
Sbjct 199 DITQIKHINNISVKLPHISDGLIFTPV 225
> cpv:cgd6_2380 mRNA capping enzyme; RNA guanylyltransferase Ceg1p
; K00987 mRNA guanylyltransferase [EC:2.7.7.50]
Length=442
Score = 89.7 bits (221), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 53/129 (41%), Positives = 75/129 (58%), Gaps = 20/129 (15%)
Query 41 QEETLLDGELVLDQLEGAA----VWRFLVYDCISIDGNETIQKLNLLKRLHAVRKQVIEP 96
Q+ TLLDGELV D +E + R+L+YDCI I+ ++T++ L LL+RL +V+ P
Sbjct 139 QQNTLLDGELVKDTIEVDGQKRYILRYLIYDCICIERDDTVKSLPLLERLKLAYLKVVIP 198
Query 97 LLLLQLQQQQQQQTAAAAAAAVGDSLPLCSKPPMEIYLKDFFEIFDLNSIRCLSLRLPHP 156
+ + Q + T + P E+YLKDFFE+ ++ +I S RLPHP
Sbjct 199 ----KCKYDQNRSTISIDPT------------PFELYLKDFFEVDEVPAILNFSRRLPHP 242
Query 157 SDGIIFTPV 165
SDGIIFTPV
Sbjct 243 SDGIIFTPV 251
> bbo:BBOV_III006110 17.m07541; mRNA capping enzyme, C-terminal
domain containing protein; K00987 mRNA guanylyltransferase
[EC:2.7.7.50]
Length=409
Score = 81.6 bits (200), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 53/139 (38%), Positives = 77/139 (55%), Gaps = 25/139 (17%)
Query 31 LPRAADSSEAQEETLLDGELVLDQLEGAAVW----RFLVYDCISIDGNETIQKLNLLKRL 86
LP + +E+Q+ TLLDGE+VLD +E + RF+ YD I + ++++LN L+R+
Sbjct 102 LPVRGNLTESQQITLLDGEIVLDTVEIDGIETQQPRFMCYDGIYVQ-RRSLKELNFLERI 160
Query 87 HAVRKQVIEPLLLLQLQQQQQQQTAAAAAAAVGDSLPLCSKPPMEIYLKDFFEIFDLNSI 146
V VI+P + + Q A KPP+ IYLKDFF+I + I
Sbjct 161 SIVYTDVIQPY-----AKSIKSQNA---------------KPPLTIYLKDFFDISHITHI 200
Query 147 RCLSLRLPHPSDGIIFTPV 165
L+ +LPH SDG+IFTPV
Sbjct 201 ENLAQQLPHVSDGLIFTPV 219
> xla:399383 rngtt, xcap1b; RNA guanylyltransferase and 5'-phosphatase
(EC:2.7.7.50); K13917 mRNA-capping enzyme [EC:2.7.7.50
3.1.3.33]
Length=598
Score = 56.6 bits (135), Expect = 4e-08, Method: Composition-based stats.
Identities = 37/146 (25%), Positives = 67/146 (45%), Gaps = 20/146 (13%)
Query 23 VREISEMYLPRAADSSEAQEETLLDGELVLDQLEGAAVWRFLVYDCISIDGNETIQKLNL 82
V ++ + P D ++ TLLDGE+++D++ G V R+L+YD I +G + + +
Sbjct 320 VFHVTNLEFPFRKDLNQHLNNTLLDGEMIIDKVNGQVVPRYLIYDIIKFNG-QPVGDCDF 378
Query 83 LKRLHAVRKQVIEPLLLLQLQQQQQQQTAAAAAAAVGDSLPLCSKPPMEIYLKDFFEIFD 142
RL + K++I P + ++ +T A K P + K FF+I
Sbjct 379 NIRLACIEKEIIFP-------RHEKMKTGLIDKA----------KEPFSVRSKPFFDIHA 421
Query 143 LNSIR--CLSLRLPHPSDGIIFTPVA 166
+ + + H DG+IF P+
Sbjct 422 ARKLLEGSFAREVSHEVDGLIFQPIG 447
> dre:405803 rngtt, MGC76886, zgc:76886; RNA guanylyltransferase
and 5'-phosphatase (EC:2.7.7.50 3.1.3.33); K13917 mRNA-capping
enzyme [EC:2.7.7.50 3.1.3.33]
Length=598
Score = 55.8 bits (133), Expect = 6e-08, Method: Composition-based stats.
Identities = 38/146 (26%), Positives = 66/146 (45%), Gaps = 20/146 (13%)
Query 23 VREISEMYLPRAADSSEAQEETLLDGELVLDQLEGAAVWRFLVYDCISIDGNETIQKLNL 82
V I + P D TLLDGE+++D++ G V R+L+YD I G + + + +
Sbjct 323 VFHIENLEFPFRKDLRIHLSNTLLDGEMIIDKVNGQPVPRYLIYDIIKFSG-QPVGQCDF 381
Query 83 LKRLHAVRKQVIEPLLLLQLQQQQQQQTAAAAAAAVGDSLPLCSKPPMEIYLKDFFEIFD 142
+RL + K++I P + ++ + Q A K P + K FF+I
Sbjct 382 NRRLLCIEKEIISP----RFEKMKLGQIDKA-------------KEPFSVRNKPFFDIHA 424
Query 143 LNSIR--CLSLRLPHPSDGIIFTPVA 166
+ + ++ H DG+IF P+
Sbjct 425 ARKLLEGSFTSQVSHEVDGLIFQPIG 450
> mmu:24018 Rngtt, AU020997; RNA guanylyltransferase and 5'-phosphatase
(EC:2.7.7.50 3.1.3.33); K13917 mRNA-capping enzyme
[EC:2.7.7.50 3.1.3.33]
Length=597
Score = 53.1 bits (126), Expect = 4e-07, Method: Composition-based stats.
Identities = 38/149 (25%), Positives = 67/149 (44%), Gaps = 26/149 (17%)
Query 23 VREISEMYLPRAADSSEAQEETLLDGELVLDQLEGAAVWRFLVYDCISIDGNETIQKLNL 82
V +S + P D TLLDGE+++D++ G AV R+L+YD I + + + +
Sbjct 319 VFHVSNLEFPFRKDLRMHLSNTLLDGEMIIDKVNGQAVPRYLIYDIIKFNA-QPVGDCDF 377
Query 83 LKRLHAVRKQVIEPLLLLQLQQQQQQQTAAAAAAAVGDSLPLCSKPPMEIYLKDFFEIFD 142
RL + +++I P + ++ +T ++ P + K F FD
Sbjct 378 NIRLQCIEREIISP-------RHEKMKTGLIDK----------TQEPFSVRPKQF---FD 417
Query 143 LNSIRCL-----SLRLPHPSDGIIFTPVA 166
+N R L + + H DG+IF P+
Sbjct 418 INISRKLLEGNFAKEVSHEMDGLIFQPIG 446
> hsa:8732 RNGTT, CAP1A, DKFZp686J2031, HCE, HCE1, hCAP; RNA guanylyltransferase
and 5'-phosphatase (EC:2.7.7.50 3.1.3.33);
K13917 mRNA-capping enzyme [EC:2.7.7.50 3.1.3.33]
Length=597
Score = 52.0 bits (123), Expect = 1e-06, Method: Composition-based stats.
Identities = 38/153 (24%), Positives = 66/153 (43%), Gaps = 34/153 (22%)
Query 23 VREISEMYLPRAADSSEAQEETLLDGELVLDQLEGAAVWRFLVYDCISIDGNETIQKLNL 82
V +S + P D TLLDGE+++D++ G AV R+L+YD I + ++ + +
Sbjct 319 VFHVSNLEFPFRKDLRMHLSNTLLDGEMIIDRVNGQAVPRYLIYDIIKFN-SQPVGDCDF 377
Query 83 LKRLHAVRKQVIEPLLLLQLQQQQQQQTAAAAAAAVGDSLPLCSKPPMEIYLKDFFEIFD 142
RL + +++I P + ++ +T ++ P + K FF+I
Sbjct 378 NVRLQCIEREIISP-------RHEKMKTGLIDK----------TQEPFSVRNKPFFDI-- 418
Query 143 LNSIRCLSLRL---------PHPSDGIIFTPVA 166
C S +L H DG+IF P
Sbjct 419 -----CTSRKLLEGNFAKEVSHEMDGLIFQPTG 446
> pfa:PF14_0144 pgt1; RNA guanylyltransferase; K00987 mRNA guanylyltransferase
[EC:2.7.7.50]
Length=520
Score = 45.4 bits (106), Expect = 1e-04, Method: Composition-based stats.
Identities = 20/37 (54%), Positives = 26/37 (70%), Gaps = 0/37 (0%)
Query 129 PMEIYLKDFFEIFDLNSIRCLSLRLPHPSDGIIFTPV 165
P EIYLKDF+ I + + + +LPH SDGIIFTP+
Sbjct 331 PFEIYLKDFYPIEKICELIKIMKKLPHYSDGIIFTPL 367
Score = 42.4 bits (98), Expect = 8e-04, Method: Composition-based stats.
Identities = 36/98 (36%), Positives = 52/98 (53%), Gaps = 12/98 (12%)
Query 11 YYLFI---YLFVYLQVREI--SEMYLPRAADSSEAQEETLLDGELVLDQL--EGAAVWR- 62
Y+LFI F+ + EI ++M++P D S+ Q+ TLLDGELV D + E V
Sbjct 68 YFLFIASNTTFLIDRNYEIFKNDMHIPTIEDLSKKQQLTLLDGELVEDIIYNEKTGVEEK 127
Query 63 ---FLVYDCISIDGNETIQKLNLLKRLHAVRKQVIEPL 97
+L+YD + I + I L+ +RL V VI PL
Sbjct 128 KIVYLIYDGLYIQ-RKDITNLSYFERLTNVYNYVITPL 164
> sce:YGL130W CEG1; Alpha (guanylyltransferase) subunit of the
mRNA capping enzyme, a heterodimer (the other subunit is CET1,
an RNA 5'-triphophatase) involved in adding the 5' cap to
mRNA; the mammalian enzyme is a single bifunctional polypeptide
(EC:2.7.7.50); K00987 mRNA guanylyltransferase [EC:2.7.7.50]
Length=459
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 38/128 (29%), Positives = 62/128 (48%), Gaps = 20/128 (15%)
Query 41 QEETLLDGELVL--DQLEGAAVWRFLVYDCISIDGNETIQKLNLLKRLHAVRKQVIEPLL 98
Q+ TLLDGELV+ + + R+L++DC++I+G + + RL + K+ +P
Sbjct 124 QDGTLLDGELVIQTNPMTKLQELRYLMFDCLAING-RCLTQSPTSSRLAHLGKEFFKPYF 182
Query 99 LLQLQQQQQQQTAAAAAAAVGDSLPLCSKPPMEIYLKDFFEIFDLNSIRCLSLRLPHPSD 158
L+ AA + C+ P +I +K + L + +LPH SD
Sbjct 183 DLR--------------AAYPNR---CTTFPFKISMKHMDFSYQLVKVAKSLDKLPHLSD 225
Query 159 GIIFTPVA 166
G+IFTPV
Sbjct 226 GLIFTPVK 233
> ath:AT5G01290 mRNA guanylyltransferase/ phosphatase/ polynucleotide
5'-phosphatase/ protein tyrosine phosphatase/ protein
tyrosine/serine/threonine phosphatase; K13917 mRNA-capping
enzyme [EC:2.7.7.50 3.1.3.33]
Length=657
Score = 41.6 bits (96), Expect = 0.001, Method: Composition-based stats.
Identities = 23/67 (34%), Positives = 38/67 (56%), Gaps = 4/67 (5%)
Query 33 RAADSSEAQEETLLDGELVLDQ---LEGAAVWRFLVYDCISIDGNETIQKLNLLKRLHAV 89
R S + TLLDGE+V+D +G A R+LVYD ++I+G +++ +R +
Sbjct 396 REGISDKVHHYTLLDGEMVIDTPTGEQGEARRRYLVYDMVAINGESVVER-TFCERWNMF 454
Query 90 RKQVIEP 96
++VI P
Sbjct 455 VREVIGP 461
> ath:AT5G28210 mRNA capping enzyme family protein; K13917 mRNA-capping
enzyme [EC:2.7.7.50 3.1.3.33]
Length=625
Score = 37.0 bits (84), Expect = 0.028, Method: Composition-based stats.
Identities = 28/85 (32%), Positives = 48/85 (56%), Gaps = 9/85 (10%)
Query 16 YLFVYLQVREISEMYLPRAADSSEAQEETLLDGELVLDQL-EGAA---VWRFLVYDCISI 71
Y F +Q+R E Y P + TLLDGE+V+D + EG V R+LVYD ++I
Sbjct 341 YRFRRVQMRFPCE-YEP---SDYKVHHYTLLDGEMVVDTIKEGETQRHVRRYLVYDLVAI 396
Query 72 DGNETIQKLNLLKRLHAVRKQVIEP 96
+G + + + +R + + +++I+P
Sbjct 397 NG-QFVAERPFSERWNILERELIKP 420
> ath:AT3G09100 mRNA capping enzyme family protein; K13917 mRNA-capping
enzyme [EC:2.7.7.50 3.1.3.33]
Length=471
Score = 34.3 bits (77), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 28/49 (57%), Gaps = 2/49 (4%)
Query 32 PRAADSSEAQEETLLDGELVLDQL--EGAAVWRFLVYDCISIDGNETIQ 78
P S + TLLDGE+++D L + R+L+YD ++I+G ++
Sbjct 412 PTEGISDKVHHFTLLDGEMIIDTLPDKQKQERRYLIYDMVAINGQSVVE 460
> mmu:80892 Zfhx4, A930021B15, C130041O22Rik, Zfh-4, Zfh4; zinc
finger homeodomain 4; K09380 zinc finger homeobox protein
4
Length=3581
Score = 31.2 bits (69), Expect = 1.6, Method: Composition-based stats.
Identities = 21/54 (38%), Positives = 28/54 (51%), Gaps = 1/54 (1%)
Query 52 LDQLEGAAVWRFLVYDCISIDGNETIQKLNLLKRLHAVRKQVIEPLLLLQLQQQ 105
L + EGA Y C D + I KLNL++ + +V+ Q E L LQL QQ
Sbjct 1005 LQKQEGAVNSESCYYYCAVCDYSSKI-KLNLVQHVRSVKHQQTEGLRKLQLHQQ 1057
> sce:YJR066W TOR1, DRR1; PIK-related protein kinase and rapamycin
target; subunit of TORC1, a complex that controls growth
in response to nutrients by regulating translation, transcription,
ribosome biogenesis, nutrient transport and autophagy;
involved in meiosis (EC:2.7.1.137); K07203 FKBP12-rapamycin
complex-associated protein
Length=2470
Score = 29.6 bits (65), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 28/58 (48%), Gaps = 1/58 (1%)
Query 82 LLKRLHAVRKQVIEPLLLLQLQQ-QQQQQTAAAAAAAVGDSLPLCSKPPMEIYLKDFF 138
L++ V K IEPLL + L + Q T A+ A L + M+IYLKD F
Sbjct 737 LIRSSKDVAKPYIEPLLNVLLPKFQDTSSTVASTALRTIGELSVVGGEDMKIYLKDLF 794
> hsa:79776 ZFHX4, FLJ16514, FLJ20980, ZFH4, ZHF4; zinc finger
homeobox 4; K09380 zinc finger homeobox protein 4
Length=3616
Score = 29.6 bits (65), Expect = 4.4, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 27/54 (50%), Gaps = 1/54 (1%)
Query 52 LDQLEGAAVWRFLVYDCISIDGNETIQKLNLLKRLHAVRKQVIEPLLLLQLQQQ 105
L + EGA Y C D + KLNL++ + +V+ Q E L LQL QQ
Sbjct 1033 LQKQEGAVNPESCYYYCAVCDYTTKV-KLNLVQHVRSVKHQQTEGLRKLQLHQQ 1085
> hsa:285679 C5orf60, FLJ35723; chromosome 5 open reading frame
60
Length=277
Score = 29.3 bits (64), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 24/101 (23%), Positives = 41/101 (40%), Gaps = 20/101 (19%)
Query 5 FICLLIYYLFIYLFVYLQVREISEMYLPRAADSSEAQEE------------TLLDGELVL 52
F+ +++ L I + + L + PR DS Q E TL D ++L
Sbjct 40 FVLFVVFSLVILIILRLYIPREPSSVPPREEDSENDQAEVGEWLRIGNKYITLKDYRILL 99
Query 53 DQLEGAAVWRFLVYDCISIDGNETIQKLNLLKRLHAVRKQV 93
+LE ++ FL C ++KL+ H + +QV
Sbjct 100 KELENLEIYTFLSKKC--------LKKLSREGSSHHLPRQV 132
> pfa:PF13_0193 MSP7-like protein
Length=298
Score = 29.3 bits (64), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 29/60 (48%), Gaps = 2/60 (3%)
Query 6 ICLLIYYLFIYLFVYLQVREISEMYLPRAADSSEAQEETLLDG-ELVLDQLEGAAVWRFL 64
I LI+YLFI FV+ V +S P D ++ LL+ E LD L+ +FL
Sbjct 2 IKGLIFYLFICFFVFF-VHAVSSQEQPNHTDIYTNEDYKLLEELEKYLDNLKNTFTHKFL 60
Lambda K H
0.326 0.141 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4027755848
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40