bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6975_orf1
Length=104
Score E
Sequences producing significant alignments: (Bits) Value
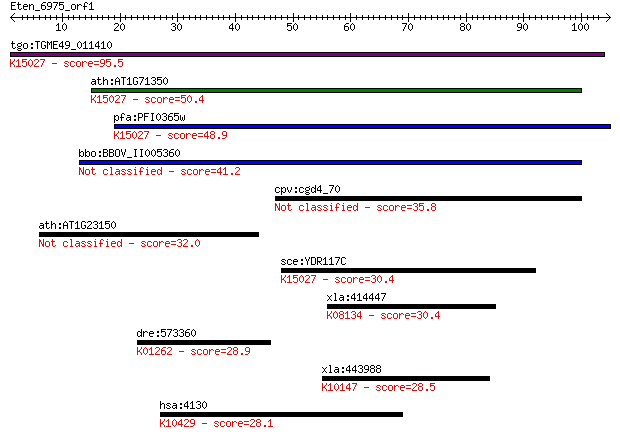
tgo:TGME49_011410 hypothetical protein ; K15027 translation in... 95.5 3e-20
ath:AT1G71350 eukaryotic translation initiation factor SUI1 fa... 50.4 1e-06
pfa:PFI0365w translation initiation factor SUI1, putative; K15... 48.9 4e-06
bbo:BBOV_II005360 18.m10002; hypothetical protein 41.2 8e-04
cpv:cgd4_70 hypothetical protein 35.8 0.036
ath:AT1G23150 hypothetical protein 32.0 0.51
sce:YDR117C TMA64; Protein of unknown function that associates... 30.4 1.5
xla:414447 lepre1, MGC84556; leucine proline-enriched proteogl... 30.4 1.5
dre:573360 xpnpep3, MGC76905, zgc:76905; X-prolyl aminopeptida... 28.9 3.7
xla:443988 ppm1d, MGC80458; protein phosphatase, Mg2+/Mn2+ dep... 28.5 4.7
hsa:4130 MAP1A, FLJ77111, MAP1L, MTAP1A; microtubule-associate... 28.1 6.3
> tgo:TGME49_011410 hypothetical protein ; K15027 translation
initiation factor 2D
Length=748
Score = 95.5 bits (236), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 48/105 (45%), Positives = 65/105 (61%), Gaps = 2/105 (1%)
Query 1 DEEAKAAGPEKLVMDKGEIFVRWMDTAQPCHAVLRDEG--ELPLLQGRIVRGACNPVRIS 58
DE A + G E+ M K E+F RW QPCH ++ L + ++ +G C PV+IS
Sbjct 598 DELATSKGSEEFSMKKEEVFSRWQAALQPCHVIVPAGAPKNLDVSTLKVHKGTCPPVKIS 657
Query 59 VEEKQGGRKHVTHIANVTNFLVEPKAVAKLLRKKLGAWASVYAPP 103
VE++ GGRKH+TH+ V FL++PK VA L+KKL A AS YA P
Sbjct 658 VEDRFGGRKHITHVVGVHVFLLDPKTVADYLQKKLAAAASTYALP 702
> ath:AT1G71350 eukaryotic translation initiation factor SUI1
family protein; K15027 translation initiation factor 2D
Length=597
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 32/85 (37%), Positives = 46/85 (54%), Gaps = 9/85 (10%)
Query 15 DKGEIFVRWMDTAQPCHAVLRDEGELPLLQGRIVRGACNPVRISVEEKQGGRKHVTHIAN 74
D G FV M QP H V+R GE P+++ +GA PV+I E +QG +K VT +
Sbjct 478 DVGSTFVGRM---QPNHVVMRGGGE-PVVR----KGAVKPVQIMTERRQGNKK-VTKVTG 528
Query 75 VTNFLVEPKAVAKLLRKKLGAWASV 99
+ FL++P + L+KK SV
Sbjct 529 METFLIDPDSFGSELQKKFACSTSV 553
> pfa:PFI0365w translation initiation factor SUI1, putative; K15027
translation initiation factor 2D
Length=818
Score = 48.9 bits (115), Expect = 4e-06, Method: Composition-based stats.
Identities = 23/86 (26%), Positives = 45/86 (52%), Gaps = 0/86 (0%)
Query 19 IFVRWMDTAQPCHAVLRDEGELPLLQGRIVRGACNPVRISVEEKQGGRKHVTHIANVTNF 78
I +++ QPC+A+++ + +I +G C + I + G+K+VTHI N+ F
Sbjct 696 IMNQFISLQQPCYAIIKPNANFDIEPIKITKGVCPSIHIYSVARMKGKKYVTHITNLYLF 755
Query 79 LVEPKAVAKLLRKKLGAWASVYAPPN 104
V+ ++ L+K+L S+ P+
Sbjct 756 HVDLNKFSEHLQKQLACSCSIVISPS 781
> bbo:BBOV_II005360 18.m10002; hypothetical protein
Length=465
Score = 41.2 bits (95), Expect = 8e-04, Method: Composition-based stats.
Identities = 28/89 (31%), Positives = 47/89 (52%), Gaps = 8/89 (8%)
Query 13 VMDKGEIFVRWMDTAQPCHAVLRDEGELPLLQGRIVRGACNPVRISVEEKQGGRKHVTHI 72
V +G + ++ ++ A H + E E P I +GA PV + VE + G RKHVT +
Sbjct 349 VSSEGPVILQHVNDAVQYHQITHGENESP-----IKKGAALPVSVLVEPR-GNRKHVTTV 402
Query 73 ANVTNFL--VEPKAVAKLLRKKLGAWASV 99
+ +L ++ VA++LR+K + SV
Sbjct 403 KGLFTYLFDLDESQVAEILRRKFASSVSV 431
> cpv:cgd4_70 hypothetical protein
Length=718
Score = 35.8 bits (81), Expect = 0.036, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 32/54 (59%), Gaps = 1/54 (1%)
Query 47 IVRGACNPVRISVEEKQGGRKHVTHIAN-VTNFLVEPKAVAKLLRKKLGAWASV 99
I +G C + I E + G RKHVT I+ +++F ++ + VA+ +KK ASV
Sbjct 623 IHKGPCKIIEIYTESRMGTRKHVTIISPFISHFNLDLQEVAESCQKKFACSASV 676
> ath:AT1G23150 hypothetical protein
Length=141
Score = 32.0 bits (71), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 24/38 (63%), Gaps = 3/38 (7%)
Query 6 AAGPEKLVMDKGEIFVRWMDTAQPCHAVLRDEGELPLL 43
+AGP + V ++G++ +DTA C+A EG LP+L
Sbjct 25 SAGPIRFVANEGDLVASVIDTALKCYA---REGRLPIL 59
> sce:YDR117C TMA64; Protein of unknown function that associates
with ribosomes; has a putative RNA binding domain; K15027
translation initiation factor 2D
Length=565
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 29/44 (65%), Gaps = 1/44 (2%)
Query 48 VRGACNPVRISVEEKQGGRKHVTHIANVTNFLVEPKAVAKLLRK 91
++G+ ++I + E + GRK +T ++N F V+P+++A LRK
Sbjct 468 MKGSLPHIKI-ITEMKIGRKVITRVSNFEVFQVDPESLAADLRK 510
> xla:414447 lepre1, MGC84556; leucine proline-enriched proteoglycan
(leprecan) 1; K08134 leucine proline-enriched proteoglycan
(leprecan)
Length=765
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 56 RISVEEKQGGRKHVTHIANVTNFLVEPKA 84
R +VEEKQ GRK ++H +V N ++ +A
Sbjct 558 RTAVEEKQDGRKDLSHPVHVDNCILNAEA 586
> dre:573360 xpnpep3, MGC76905, zgc:76905; X-prolyl aminopeptidase
(aminopeptidase P) 3, putative (EC:3.4.11.9); K01262 Xaa-Pro
aminopeptidase [EC:3.4.11.9]
Length=510
Score = 28.9 bits (63), Expect = 3.7, Method: Composition-based stats.
Identities = 11/23 (47%), Positives = 13/23 (56%), Gaps = 0/23 (0%)
Query 23 WMDTAQPCHAVLRDEGELPLLQG 45
W D +QPCH L PLL+G
Sbjct 207 WYDNSQPCHQRLHQTHVRPLLEG 229
> xla:443988 ppm1d, MGC80458; protein phosphatase, Mg2+/Mn2+ dependent,
1D (EC:3.1.3.16); K10147 protein phosphatase 1D (formerly
2C) [EC:3.1.3.16]
Length=554
Score = 28.5 bits (62), Expect = 4.7, Method: Composition-based stats.
Identities = 14/29 (48%), Positives = 20/29 (68%), Gaps = 3/29 (10%)
Query 55 VRISVEEKQGGRKHVTHIANVTNFLVEPK 83
+R+SV QGGRK ++ +VT LVEP+
Sbjct 7 LRVSVSSDQGGRK---YMEDVTQILVEPE 32
> hsa:4130 MAP1A, FLJ77111, MAP1L, MTAP1A; microtubule-associated
protein 1A; K10429 microtubule-associated protein 1
Length=2803
Score = 28.1 bits (61), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 22/42 (52%), Gaps = 2/42 (4%)
Query 27 AQPCHAVLRDEGELPLLQGRIVRGACNPVRISVEEKQGGRKH 68
A P A + +G P L G I+ +C+P R S K+ GR H
Sbjct 2061 AVPPRAPILSKGPSPPLNGNIL--SCSPDRRSPSPKESGRSH 2100
Lambda K H
0.318 0.135 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2022291452
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40