bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6983_orf1
Length=144
Score E
Sequences producing significant alignments: (Bits) Value
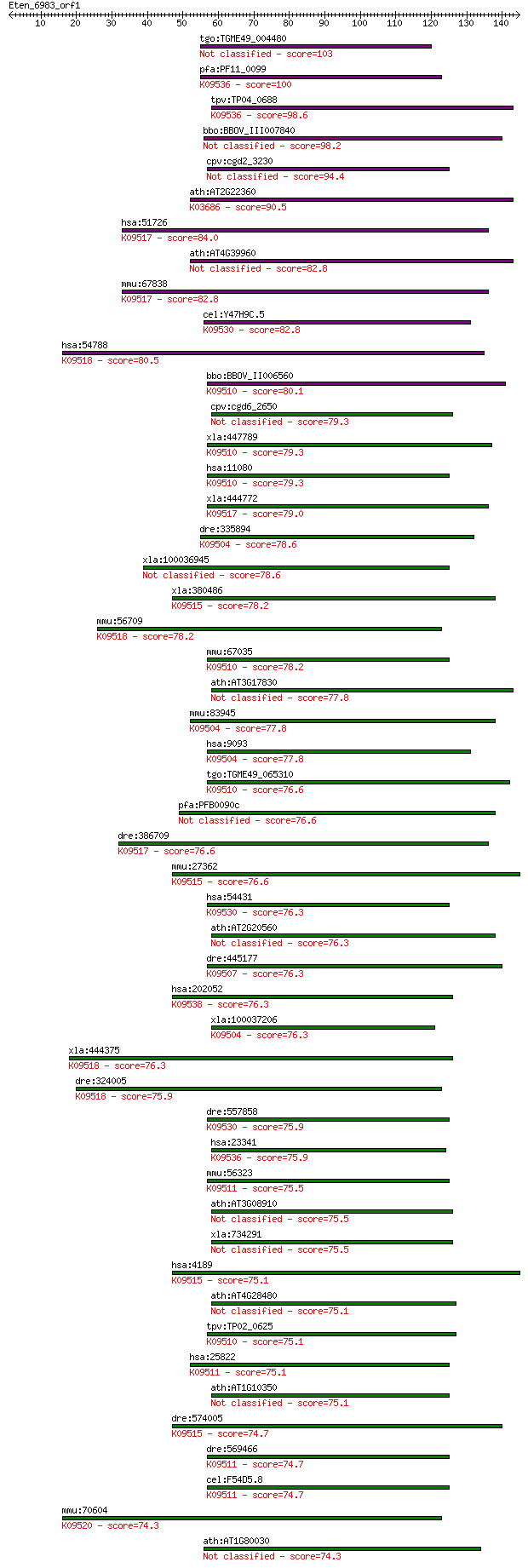
tgo:TGME49_004480 DnaJ domain-containing protein (EC:5.3.4.1) 103 2e-22
pfa:PF11_0099 heat shock protein DnaJ homologue Pfj2; K09536 D... 100 2e-21
tpv:TP04_0688 heat shock protein DnaJ; K09536 DnaJ homolog sub... 98.6 5e-21
bbo:BBOV_III007840 17.m07688; dnaJ domain containing protein 98.2 8e-21
cpv:cgd2_3230 heat shock protein DnaJ Pfj2 94.4 1e-19
ath:AT2G22360 DNAJ heat shock family protein; K03686 molecular... 90.5 2e-18
hsa:51726 DNAJB11, ABBP-2, ABBP2, DJ9, EDJ, ERdj3, ERj3, HEDJ,... 84.0 1e-16
ath:AT4G39960 DNAJ heat shock family protein 82.8 3e-16
mmu:67838 Dnajb11, 1810031F23Rik, ABBP-2, AL024055, Dj9, ERdj3... 82.8 4e-16
cel:Y47H9C.5 dnj-27; DNaJ domain (prokaryotic heat shock prote... 82.8 4e-16
hsa:54788 DNAJB12, DJ10, DKFZp586B2023; DnaJ (Hsp40) homolog, ... 80.5 2e-15
bbo:BBOV_II006560 18.m06540; protein with DnaJ domain, DNJ1/SI... 80.1 2e-15
cpv:cgd6_2650 heat shock protein 79.3 4e-15
xla:447789 dnajb4, MGC83507; DnaJ (Hsp40) homolog, subfamily B... 79.3 4e-15
hsa:11080 DNAJB4, DNAJW, DjB4, HLJ1; DnaJ (Hsp40) homolog, sub... 79.3 4e-15
xla:444772 dnajb11, MGC81924; DnaJ (Hsp40) homolog, subfamily ... 79.0 5e-15
dre:335894 dnaja3a, wu:fj36c02; DnaJ (Hsp40) homolog, subfamil... 78.6 6e-15
xla:100036945 hypothetical protein LOC100036945 78.6 6e-15
xla:380486 dnajb9, MGC53411; DnaJ (Hsp40) homolog, subfamily B... 78.2 7e-15
mmu:56709 Dnajb12, Dj10, mDj10; DnaJ (Hsp40) homolog, subfamil... 78.2 7e-15
mmu:67035 Dnajb4, 1700029A20Rik, 2010306G19Rik, 5730460G06Rik;... 78.2 8e-15
ath:AT3G17830 DNAJ heat shock family protein 77.8 9e-15
mmu:83945 Dnaja3, 1200003J13Rik, 1810053A11Rik, C81173, Tid-1,... 77.8 1e-14
hsa:9093 DNAJA3, FLJ45758, TID1, hTid-1; DnaJ (Hsp40) homolog,... 77.8 1e-14
tgo:TGME49_065310 heat shock protein 40, putative ; K09510 Dna... 76.6 2e-14
pfa:PFB0090c RESA-like protein with PHIST and DnaJ domains 76.6 2e-14
dre:386709 dnajb11, cb954; DnaJ (Hsp40) homolog, subfamily B, ... 76.6 2e-14
mmu:27362 Dnajb9, AA408011, AA673251, AA673481, AW556981, ERdj... 76.6 2e-14
hsa:54431 DNAJC10, DKFZp434J1813, ERdj5, JPDI, MGC104194; DnaJ... 76.3 3e-14
ath:AT2G20560 DNAJ heat shock family protein 76.3 3e-14
dre:445177 dnajb1a, dnajb1, hspf1, zf-Hsp40, zgc:101068; DnaJ ... 76.3 3e-14
hsa:202052 DNAJC18, MGC29463; DnaJ (Hsp40) homolog, subfamily ... 76.3 3e-14
xla:100037206 dnaja3, tid1; DnaJ (Hsp40) homolog, subfamily A,... 76.3 3e-14
xla:444375 dnajb12, MGC82876; DnaJ (Hsp40) homolog, subfamily ... 76.3 4e-14
dre:324005 dnajb12, wu:fc16f06, wu:fi38b10; DnaJ (Hsp40) homol... 75.9 4e-14
dre:557858 dnajc10, MGC162218, zgc:162218; DnaJ (Hsp40) homolo... 75.9 4e-14
hsa:23341 DNAJC16, DKFZp686G1298, DKFZp686N0387, DKFZp781I1547... 75.9 5e-14
mmu:56323 Dnajb5, 1110058L06Rik, AI462558, Hsc40, Hsp40-3; Dna... 75.5 5e-14
ath:AT3G08910 DNAJ heat shock protein, putative 75.5 5e-14
xla:734291 dnajb6-a, MGC85133; dnaJ homolog subfamily B member... 75.5 6e-14
hsa:4189 DNAJB9, DKFZp564F1862, ERdj4, MDG-1, MDG1, MST049, MS... 75.1 7e-14
ath:AT4G28480 DNAJ heat shock family protein 75.1 7e-14
tpv:TP02_0625 chaperone protein DnaJ; K09510 DnaJ homolog subf... 75.1 7e-14
hsa:25822 DNAJB5, Hsc40, KIAA1045; DnaJ (Hsp40) homolog, subfa... 75.1 8e-14
ath:AT1G10350 DNAJ heat shock protein, putative 75.1 8e-14
dre:574005 zgc:114162; K09515 DnaJ homolog subfamily B member 9 74.7
dre:569466 novel protein similar to vertebrate DnaJ (Hsp40) ho... 74.7 9e-14
cel:F54D5.8 dnj-13; DNaJ domain (prokaryotic heat shock protei... 74.7 1e-13
mmu:70604 Dnajb14, 5730496F10Rik; DnaJ (Hsp40) homolog, subfam... 74.3 1e-13
ath:AT1G80030 DNAJ heat shock protein, putative 74.3 1e-13
> tgo:TGME49_004480 DnaJ domain-containing protein (EC:5.3.4.1)
Length=606
Score = 103 bits (257), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 47/65 (72%), Positives = 58/65 (89%), Gaps = 0/65 (0%)
Query 55 KSKDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVAPGKEKEFMEIAKAHEVLSDPEQRK 114
KS+D+Y+LLGV+RNA+ R IDKAYRKLAK+ HPDV PG E++F++IAKAHEVLS+ EQRK
Sbjct 68 KSRDFYRLLGVKRNATPREIDKAYRKLAKQYHPDVNPGNEEKFLDIAKAHEVLSNEEQRK 127
Query 115 KYDTF 119
KYD F
Sbjct 128 KYDMF 132
> pfa:PF11_0099 heat shock protein DnaJ homologue Pfj2; K09536
DnaJ homolog subfamily C member 16
Length=540
Score = 100 bits (249), Expect = 2e-21, Method: Composition-based stats.
Identities = 42/68 (61%), Positives = 56/68 (82%), Gaps = 0/68 (0%)
Query 55 KSKDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVAPGKEKEFMEIAKAHEVLSDPEQRK 114
+ DYY+ LGV+RNA++ I KAYR+LAK+ HPD+AP KEK+F+EIA A+E LSDPE+RK
Sbjct 32 RGMDYYKRLGVKRNATKEDISKAYRQLAKEYHPDIAPDKEKDFIEIANAYETLSDPEKRK 91
Query 115 KYDTFGEE 122
YD +GE+
Sbjct 92 MYDMYGED 99
> tpv:TP04_0688 heat shock protein DnaJ; K09536 DnaJ homolog subfamily
C member 16
Length=509
Score = 98.6 bits (244), Expect = 5e-21, Method: Composition-based stats.
Identities = 45/85 (52%), Positives = 57/85 (67%), Gaps = 5/85 (5%)
Query 58 DYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVAPGKEKEFMEIAKAHEVLSDPEQRKKYD 117
DYY +LGV++NA+ R I+KA+RK AKKLHPD PG EK F E++ A+EVL DP +R+ YD
Sbjct 23 DYYSILGVKKNATDREIEKAFRKKAKKLHPDANPGNEKAFAELSNAYEVLKDPSKRQTYD 82
Query 118 TFGEEGLAGFPGAGAGDAGFPFSEY 142
GEEGL G G P+ Y
Sbjct 83 MHGEEGL-----KQEGPQGHPYQHY 102
> bbo:BBOV_III007840 17.m07688; dnaJ domain containing protein
Length=387
Score = 98.2 bits (243), Expect = 8e-21, Method: Compositional matrix adjust.
Identities = 50/84 (59%), Positives = 58/84 (69%), Gaps = 0/84 (0%)
Query 56 SKDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVAPGKEKEFMEIAKAHEVLSDPEQRKK 115
+KDYY LLGV RNAS I KAYR AKKLHPDVAPGKE+EF +I A+EVL D E+RK+
Sbjct 32 AKDYYSLLGVSRNASDADIAKAYRSKAKKLHPDVAPGKEEEFKDINTAYEVLKDSEKRKQ 91
Query 116 YDTFGEEGLAGFPGAGAGDAGFPF 139
YD +GE G+ G G G F
Sbjct 92 YDLYGEAGVNGAGAQSQGQQGHDF 115
> cpv:cgd2_3230 heat shock protein DnaJ Pfj2
Length=601
Score = 94.4 bits (233), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 42/68 (61%), Positives = 55/68 (80%), Gaps = 0/68 (0%)
Query 57 KDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVAPGKEKEFMEIAKAHEVLSDPEQRKKY 116
KDYY++LGV RNA++ I +AYRKL+ K HPD PG +++FME+A A+EVL +PE R+KY
Sbjct 22 KDYYKILGVPRNANENQIKRAYRKLSLKYHPDKNPGSKEKFMEVANAYEVLVNPETRRKY 81
Query 117 DTFGEEGL 124
D FGEEGL
Sbjct 82 DAFGEEGL 89
> ath:AT2G22360 DNAJ heat shock family protein; K03686 molecular
chaperone DnaJ
Length=442
Score = 90.5 bits (223), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 46/93 (49%), Positives = 59/93 (63%), Gaps = 2/93 (2%)
Query 52 AARKSKDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVA--PGKEKEFMEIAKAHEVLSD 109
R DYY +LGV +NA++ I AYRKLA+ HPDV PG E++F EI+ A+EVLSD
Sbjct 80 TVRADADYYSVLGVSKNATKAEIKSAYRKLARNYHPDVNKDPGAEEKFKEISNAYEVLSD 139
Query 110 PEQRKKYDTFGEEGLAGFPGAGAGDAGFPFSEY 142
E++ YD +GE GL G G G GD PF +
Sbjct 140 DEKKSLYDRYGEAGLKGAAGFGNGDFSNPFDLF 172
> hsa:51726 DNAJB11, ABBP-2, ABBP2, DJ9, EDJ, ERdj3, ERj3, HEDJ,
PRO1080, UNQ537, hDj9; DnaJ (Hsp40) homolog, subfamily B,
member 11; K09517 DnaJ homolog subfamily B member 11
Length=358
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 48/106 (45%), Positives = 67/106 (63%), Gaps = 5/106 (4%)
Query 33 APHSSGGFWSSYGGIIGAEAARKSKDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVAPG 92
AP + F +IGA A +D+Y++LGV R+AS + I KAYRKLA +LHPD P
Sbjct 2 APQNLSTFCLLLLYLIGAVIA--GRDFYKILGVPRSASIKDIKKAYRKLALQLHPDRNPD 59
Query 93 K---EKEFMEIAKAHEVLSDPEQRKKYDTFGEEGLAGFPGAGAGDA 135
+++F ++ A+EVLSD E+RK+YDT+GEEGL + GD
Sbjct 60 DPQAQEKFQDLGAAYEVLSDSEKRKQYDTYGEEGLKDGHQSSHGDI 105
> ath:AT4G39960 DNAJ heat shock family protein
Length=447
Score = 82.8 bits (203), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 42/93 (45%), Positives = 56/93 (60%), Gaps = 2/93 (2%)
Query 52 AARKSKDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVA--PGKEKEFMEIAKAHEVLSD 109
R D+Y +LGV +NA++ I AYRKLA+ HPDV G E +F EI+ A+E+LSD
Sbjct 79 TVRADTDFYSVLGVSKNATKAEIKSAYRKLARSYHPDVNKDAGAEDKFKEISNAYEILSD 138
Query 110 PEQRKKYDTFGEEGLAGFPGAGAGDAGFPFSEY 142
E+R YD +GE G+ G G GD PF +
Sbjct 139 DEKRSLYDRYGEAGVKGAGMGGMGDYSNPFDLF 171
> mmu:67838 Dnajb11, 1810031F23Rik, ABBP-2, AL024055, Dj9, ERdj3,
ERj3p; DnaJ (Hsp40) homolog, subfamily B, member 11; K09517
DnaJ homolog subfamily B member 11
Length=358
Score = 82.8 bits (203), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 47/106 (44%), Positives = 66/106 (62%), Gaps = 5/106 (4%)
Query 33 APHSSGGFWSSYGGIIGAEAARKSKDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVAPG 92
AP + F +IG A +D+Y++LGV R+AS + I KAYRKLA +LHPD P
Sbjct 2 APQNLSTFCLLLLYLIGTVIA--GRDFYKILGVPRSASIKDIKKAYRKLALQLHPDRNPD 59
Query 93 K---EKEFMEIAKAHEVLSDPEQRKKYDTFGEEGLAGFPGAGAGDA 135
+++F ++ A+EVLSD E+RK+YDT+GEEGL + GD
Sbjct 60 DPQAQEKFQDLGAAYEVLSDSEKRKQYDTYGEEGLKDGHQSSHGDI 105
> cel:Y47H9C.5 dnj-27; DNaJ domain (prokaryotic heat shock protein)
family member (dnj-27); K09530 DnaJ homolog subfamily
C member 10
Length=788
Score = 82.8 bits (203), Expect = 4e-16, Method: Composition-based stats.
Identities = 43/79 (54%), Positives = 53/79 (67%), Gaps = 4/79 (5%)
Query 56 SKDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVA---PGKEKEFMEIAKAHEVLSDPEQ 112
++DYY+LLGV R+A R I KA++KLA K HPD P EF++I KA+EVL D
Sbjct 18 AEDYYELLGVERDADDRTIRKAFKKLAIKKHPDRNTDDPNAHDEFVKINKAYEVLKDENL 77
Query 113 RKKYDTFGEEGLA-GFPGA 130
RKKYD FGE+GL GF G
Sbjct 78 RKKYDQFGEKGLEDGFQGG 96
> hsa:54788 DNAJB12, DJ10, DKFZp586B2023; DnaJ (Hsp40) homolog,
subfamily B, member 12; K09518 DnaJ homolog subfamily B member
12
Length=409
Score = 80.5 bits (197), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 51/128 (39%), Positives = 73/128 (57%), Gaps = 9/128 (7%)
Query 16 PPNPSSRSSTNSGGPYGAPHSSG--GFWSSYG----GIIGAEAARKSKDYYQLLGVRRNA 69
PP ++ ++ G AP ++G G S+ G + + ++ KDYY++LGV R A
Sbjct 96 PPTDTTHATHRKAGGTDAPSANGEAGGESTKGYTAEQVAAVKRVKQCKDYYEILGVSRGA 155
Query 70 SQRAIDKAYRKLAKKLHPDV--APGKEKEFMEIAKAHEVLSDPEQRKKYDTFGEE-GLAG 126
S + KAYR+LA K HPD APG + F I A+ VLS+PE+RK+YD FG++ A
Sbjct 156 SDEDLKKAYRRLALKFHPDKNHAPGATEAFKAIGTAYAVLSNPEKRKQYDQFGDDKSQAA 215
Query 127 FPGAGAGD 134
G G GD
Sbjct 216 RHGHGHGD 223
> bbo:BBOV_II006560 18.m06540; protein with DnaJ domain, DNJ1/SIS1
family; K09510 DnaJ homolog subfamily B member 4
Length=323
Score = 80.1 bits (196), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 44/93 (47%), Positives = 58/93 (62%), Gaps = 9/93 (9%)
Query 57 KDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVAPG------KEKEFMEIAKAHEVLSDP 110
KDYY +LGV R ++ + KAYRKLA + HPD P E F +++A++VLSDP
Sbjct 3 KDYYSILGVSRGSNDAELKKAYRKLAMQWHPDKHPDPVAKQKAEDMFKNVSEAYDVLSDP 62
Query 111 EQRKKYDTFGEEGL---AGFPGAGAGDAGFPFS 140
E+RK YD FGEEGL AG P GAG + ++
Sbjct 63 EKRKIYDQFGEEGLKGTAGGPNQGAGTTQYVYT 95
> cpv:cgd6_2650 heat shock protein
Length=273
Score = 79.3 bits (194), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 38/72 (52%), Positives = 53/72 (73%), Gaps = 4/72 (5%)
Query 58 DYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVAPGKEKE----FMEIAKAHEVLSDPEQR 113
DYY++L V+R+AS I K+YRKLA K HPD P +E F +IA+A+EVLSDPE+R
Sbjct 2 DYYEILEVKRDASTSEIKKSYRKLALKWHPDKNPDNREEAEEMFKKIAEAYEVLSDPEKR 61
Query 114 KKYDTFGEEGLA 125
+YDT+G +G++
Sbjct 62 NRYDTYGADGVS 73
> xla:447789 dnajb4, MGC83507; DnaJ (Hsp40) homolog, subfamily
B, member 4; K09510 DnaJ homolog subfamily B member 4
Length=339
Score = 79.3 bits (194), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 44/82 (53%), Positives = 57/82 (69%), Gaps = 2/82 (2%)
Query 57 KDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDV--APGKEKEFMEIAKAHEVLSDPEQRK 114
KDYY +LG+ + AS+ I KAYRK A K HPD + E++F EIA+A+EVLSDP++++
Sbjct 3 KDYYSILGIEKGASEDDIKKAYRKQALKWHPDKNKSAHAEEKFKEIAEAYEVLSDPKKKE 62
Query 115 KYDTFGEEGLAGFPGAGAGDAG 136
YD FGEEGL G GA G G
Sbjct 63 VYDQFGEEGLKGGSGAPDGHGG 84
> hsa:11080 DNAJB4, DNAJW, DjB4, HLJ1; DnaJ (Hsp40) homolog, subfamily
B, member 4; K09510 DnaJ homolog subfamily B member
4
Length=337
Score = 79.3 bits (194), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 40/70 (57%), Positives = 52/70 (74%), Gaps = 2/70 (2%)
Query 57 KDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDV--APGKEKEFMEIAKAHEVLSDPEQRK 114
KDYY +LG+ + AS I KAYRK A K HPD +P E++F E+A+A+EVLSDP++R+
Sbjct 3 KDYYCILGIEKGASDEDIKKAYRKQALKFHPDKNKSPQAEEKFKEVAEAYEVLSDPKKRE 62
Query 115 KYDTFGEEGL 124
YD FGEEGL
Sbjct 63 IYDQFGEEGL 72
> xla:444772 dnajb11, MGC81924; DnaJ (Hsp40) homolog, subfamily
B, member 11; K09517 DnaJ homolog subfamily B member 11
Length=360
Score = 79.0 bits (193), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 40/82 (48%), Positives = 56/82 (68%), Gaps = 3/82 (3%)
Query 57 KDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVAPGK---EKEFMEIAKAHEVLSDPEQR 113
+D+Y++LGV R A+ + I KAYRKLA +LHPD P + +F ++ A+EVLSD E+R
Sbjct 26 RDFYKILGVSRGATVKEIKKAYRKLALQLHPDRNPDDPNAQDKFQDLGAAYEVLSDEEKR 85
Query 114 KKYDTFGEEGLAGFPGAGAGDA 135
K+YDT+GEEGL + GD
Sbjct 86 KQYDTYGEEGLKDGHQSSHGDI 107
> dre:335894 dnaja3a, wu:fj36c02; DnaJ (Hsp40) homolog, subfamily
A, member 3A; K09504 DnaJ homolog subfamily A member 3
Length=453
Score = 78.6 bits (192), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 40/81 (49%), Positives = 59/81 (72%), Gaps = 4/81 (4%)
Query 55 KSKDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVA---PGKEKEFMEIAKAHEVLSDPE 111
+ +D+YQ+LGV R+A+Q+ I KAY ++AKK HPD P +++F ++A+A+EVLSD
Sbjct 88 RKQDFYQILGVPRSATQKEIKKAYYQMAKKYHPDTNKEDPQAKEKFAQLAEAYEVLSDEV 147
Query 112 QRKKYDTFGEEGL-AGFPGAG 131
+RK+YDT+G G AG GAG
Sbjct 148 KRKQYDTYGSAGFDAGRAGAG 168
> xla:100036945 hypothetical protein LOC100036945
Length=358
Score = 78.6 bits (192), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 41/89 (46%), Positives = 58/89 (65%), Gaps = 7/89 (7%)
Query 39 GFWSSYGGIIGAEAARKSKDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVAPGK---EK 95
GF Y ++ +D+Y++LGV R A+ + I KAYRKLA +LHPD P +
Sbjct 10 GFLICYLMVV----VSGGRDFYKILGVSRGATVKEIKKAYRKLALQLHPDRNPDDPNAQD 65
Query 96 EFMEIAKAHEVLSDPEQRKKYDTFGEEGL 124
+F ++ A+EVLSD E+RK+YDT+GEEGL
Sbjct 66 KFQDLGAAYEVLSDEEKRKQYDTYGEEGL 94
> xla:380486 dnajb9, MGC53411; DnaJ (Hsp40) homolog, subfamily
B, member 9; K09515 DnaJ homolog subfamily B member 9
Length=221
Score = 78.2 bits (191), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 43/93 (46%), Positives = 57/93 (61%), Gaps = 3/93 (3%)
Query 47 IIGAEAARKSKDYYQLLGVRRNASQRAIDKAYRKLAKKLHPD--VAPGKEKEFMEIAKAH 104
++ +E K YY +LGV +NAS+R I KA+ KLA K HPD +P E +F EIA+A+
Sbjct 15 LLISEIILAKKTYYDILGVPKNASERQIKKAFHKLAMKYHPDKNKSPDAETKFREIAEAY 74
Query 105 EVLSDPEQRKKYDTFGEEGLAGFPGAGAGDAGF 137
E LSD +RK+YD FG + A G G D F
Sbjct 75 ETLSDESKRKEYDQFGHDAFAN-GGGGGSDQHF 106
> mmu:56709 Dnajb12, Dj10, mDj10; DnaJ (Hsp40) homolog, subfamily
B, member 12; K09518 DnaJ homolog subfamily B member 12
Length=376
Score = 78.2 bits (191), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 43/99 (43%), Positives = 62/99 (62%), Gaps = 4/99 (4%)
Query 26 NSGGPYGAPHSSGGFWSSYGGIIGAEAARKSKDYYQLLGVRRNASQRAIDKAYRKLAKKL 85
++ G G S+ G+ S + + ++ KDYY++LGV R+AS + KAYRKLA K
Sbjct 81 SANGEAGGGESAKGYTSE--QVAAVKRVKQCKDYYEILGVSRSASDEDLKKAYRKLALKF 138
Query 86 HPDV--APGKEKEFMEIAKAHEVLSDPEQRKKYDTFGEE 122
HPD APG + F I A+ VLS+PE+RK+YD FG++
Sbjct 139 HPDKNHAPGATEAFKAIGTAYAVLSNPEKRKQYDQFGDD 177
> mmu:67035 Dnajb4, 1700029A20Rik, 2010306G19Rik, 5730460G06Rik;
DnaJ (Hsp40) homolog, subfamily B, member 4; K09510 DnaJ
homolog subfamily B member 4
Length=337
Score = 78.2 bits (191), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 38/70 (54%), Positives = 52/70 (74%), Gaps = 2/70 (2%)
Query 57 KDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDV--APGKEKEFMEIAKAHEVLSDPEQRK 114
KDYY +LG+ + A+ + KAYRK A K HPD +P E++F E+A+A+EVLSDP++R+
Sbjct 3 KDYYHILGIDKGATDEDVKKAYRKQALKFHPDKNKSPQAEEKFKEVAEAYEVLSDPKKRE 62
Query 115 KYDTFGEEGL 124
YD FGEEGL
Sbjct 63 IYDQFGEEGL 72
> ath:AT3G17830 DNAJ heat shock family protein
Length=517
Score = 77.8 bits (190), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 44/89 (49%), Positives = 56/89 (62%), Gaps = 4/89 (4%)
Query 58 DYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVA--PGKEKEFMEIAKAHEVLSDPEQRKK 115
D+Y L V RNA+ + I +YRKLA+K HPD+ PG E +F +I+ A+EVLSD E+R
Sbjct 63 DHYSTLNVNRNATLQEIKSSYRKLARKYHPDMNKNPGAEDKFKQISAAYEVLSDEEKRSA 122
Query 116 YDTFGEEGLAG-FPGAGAGDAGF-PFSEY 142
YD FGE GL G F G+ G PF Y
Sbjct 123 YDRFGEAGLEGDFNGSQDTSPGVDPFDLY 151
> mmu:83945 Dnaja3, 1200003J13Rik, 1810053A11Rik, C81173, Tid-1,
Tid1l; DnaJ (Hsp40) homolog, subfamily A, member 3; K09504
DnaJ homolog subfamily A member 3
Length=453
Score = 77.8 bits (190), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 42/89 (47%), Positives = 58/89 (65%), Gaps = 5/89 (5%)
Query 52 AARKSKDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVA---PGKEKEFMEIAKAHEVLS 108
A+ DYYQ+LGV RNASQ+ I KAY +LAKK HPD P +++F ++A+A+EVLS
Sbjct 87 ASLAKDDYYQILGVPRNASQKDIKKAYYQLAKKYHPDTNKDDPKAKEKFSQLAEAYEVLS 146
Query 109 DPEQRKKYDTFGEEGLAGFPGAGAGDAGF 137
D +RK+YD +G G PG + G+
Sbjct 147 DEVKRKQYDAYGSAGFD--PGTSSSGQGY 173
> hsa:9093 DNAJA3, FLJ45758, TID1, hTid-1; DnaJ (Hsp40) homolog,
subfamily A, member 3; K09504 DnaJ homolog subfamily A member
3
Length=453
Score = 77.8 bits (190), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 41/77 (53%), Positives = 55/77 (71%), Gaps = 5/77 (6%)
Query 57 KDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVA---PGKEKEFMEIAKAHEVLSDPEQR 113
+DYYQ+LGV RNASQ+ I KAY +LAKK HPD P +++F ++A+A+EVLSD +R
Sbjct 92 EDYYQILGVPRNASQKEIKKAYYQLAKKYHPDTNKDDPKAKEKFSQLAEAYEVLSDEVKR 151
Query 114 KKYDTFGEEGLAGFPGA 130
K+YD +G G PGA
Sbjct 152 KQYDAYGSAGFD--PGA 166
> tgo:TGME49_065310 heat shock protein 40, putative ; K09510 DnaJ
homolog subfamily B member 4
Length=336
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 47/97 (48%), Positives = 62/97 (63%), Gaps = 12/97 (12%)
Query 57 KDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDV---APGKEK---EFMEIAKAHEVLSDP 110
KDYY++LGV ++AS+ + KAYRKLA K HPD A K+K +F +IA+A++VLSD
Sbjct 3 KDYYRILGVGKDASEADLKKAYRKLAMKWHPDKHADADAKKKAEAQFKDIAEAYDVLSDK 62
Query 111 EQRKKYDTFGEEGL------AGFPGAGAGDAGFPFSE 141
E+R+ YD FGEEGL G G G A F + E
Sbjct 63 EKRQIYDQFGEEGLKSGGSPTGTAGPGGSRANFVYRE 99
> pfa:PFB0090c RESA-like protein with PHIST and DnaJ domains
Length=421
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 42/95 (44%), Positives = 57/95 (60%), Gaps = 6/95 (6%)
Query 49 GAEAARKSKDYYQLLGVRRNASQRAIDKAYRKLAKKLHPD----VAPGKEKE--FMEIAK 102
G + +K++DYY +LGV R+ + I KAY+KLA K HPD A KE + F I++
Sbjct 79 GKASTKKNEDYYSILGVSRDCTNEDIKKAYKKLAMKWHPDKHLNAASKKEADNMFKSISE 138
Query 103 AHEVLSDPEQRKKYDTFGEEGLAGFPGAGAGDAGF 137
A+EVLSD E+R YD +GEEGL + GF
Sbjct 139 AYEVLSDEEKRDIYDKYGEEGLDKYGSNNGHSKGF 173
> dre:386709 dnajb11, cb954; DnaJ (Hsp40) homolog, subfamily B,
member 11; K09517 DnaJ homolog subfamily B member 11
Length=360
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 48/107 (44%), Positives = 64/107 (59%), Gaps = 7/107 (6%)
Query 32 GAPHSSGGFWSSYGGIIGAEAARKSKDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVA- 90
G SS F Y I+ A R D+Y++LGV R+AS + I KAYRKLA +LHPD
Sbjct 5 GMKLSSVCFLLLYL-ILTVFAGR---DFYKILGVSRSASVKDIKKAYRKLALQLHPDRNQ 60
Query 91 --PGKEKEFMEIAKAHEVLSDPEQRKKYDTFGEEGLAGFPGAGAGDA 135
P + +F ++ A+EVLSD E+RK+YD +GEEGL + GD
Sbjct 61 DDPNAQDKFADLGAAYEVLSDEEKRKQYDAYGEEGLKEGHQSSHGDI 107
> mmu:27362 Dnajb9, AA408011, AA673251, AA673481, AW556981, ERdj4,
Mdg1, mDj7; DnaJ (Hsp40) homolog, subfamily B, member 9;
K09515 DnaJ homolog subfamily B member 9
Length=222
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 44/100 (44%), Positives = 57/100 (57%), Gaps = 4/100 (4%)
Query 47 IIGAEAARKSKDYYQLLGVRRNASQRAIDKAYRKLAKKLHPD--VAPGKEKEFMEIAKAH 104
++ E SK YY +LGV ++AS+R I KA+ KLA K HPD +P E +F EIA+A+
Sbjct 15 LMITELILASKSYYDILGVPKSASERQIKKAFHKLAMKYHPDKNKSPDAEAKFREIAEAY 74
Query 105 EVLSDPEQRKKYDTFGEEGLAGFPGAGAGDAGFPFSEYVN 144
E LSD RK+YDT G G G G PF + N
Sbjct 75 ETLSDANSRKEYDTIGHSAFTN--GKGQRGNGSPFEQSFN 112
> hsa:54431 DNAJC10, DKFZp434J1813, ERdj5, JPDI, MGC104194; DnaJ
(Hsp40) homolog, subfamily C, member 10; K09530 DnaJ homolog
subfamily C member 10
Length=793
Score = 76.3 bits (186), Expect = 3e-14, Method: Composition-based stats.
Identities = 36/71 (50%), Positives = 50/71 (70%), Gaps = 3/71 (4%)
Query 57 KDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVAPGKEK---EFMEIAKAHEVLSDPEQR 113
+D+Y LLGV + AS R I +A++KLA KLHPD P +F++I +A+EVL D + R
Sbjct 34 QDFYSLLGVSKTASSREIRQAFKKLALKLHPDKNPNNPNAHGDFLKINRAYEVLKDEDLR 93
Query 114 KKYDTFGEEGL 124
KKYD +GE+GL
Sbjct 94 KKYDKYGEKGL 104
> ath:AT2G20560 DNAJ heat shock family protein
Length=337
Score = 76.3 bits (186), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 42/85 (49%), Positives = 57/85 (67%), Gaps = 5/85 (5%)
Query 58 DYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVAPGKEKE----FMEIAKAHEVLSDPEQR 113
DYY++L V R+AS + KAYRKLA K HPD P +K+ F +I++A+EVLSDP+++
Sbjct 4 DYYKVLQVDRSASDDDLKKAYRKLAMKWHPDKNPNNKKDAEAMFKQISEAYEVLSDPQKK 63
Query 114 KKYDTFGEEGLAG-FPGAGAGDAGF 137
YD +GEEGL G P AG A +
Sbjct 64 AVYDQYGEEGLKGNVPPPDAGGATY 88
> dre:445177 dnajb1a, dnajb1, hspf1, zf-Hsp40, zgc:101068; DnaJ
(Hsp40) homolog, subfamily B, member 1a; K09507 DnaJ homolog
subfamily B member 1
Length=335
Score = 76.3 bits (186), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 39/85 (45%), Positives = 56/85 (65%), Gaps = 2/85 (2%)
Query 57 KDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDV--APGKEKEFMEIAKAHEVLSDPEQRK 114
KDYY++LG+ + AS I KAYRK A + HPD + G E +F EIA+A++VLSD +++
Sbjct 3 KDYYRILGIEKGASDEEIKKAYRKQALRFHPDKNKSAGAEDKFKEIAEAYDVLSDAKKKD 62
Query 115 KYDTFGEEGLAGFPGAGAGDAGFPF 139
YD +GE+GL G G+G + F
Sbjct 63 IYDRYGEDGLKGHAGSGTNGPSYTF 87
> hsa:202052 DNAJC18, MGC29463; DnaJ (Hsp40) homolog, subfamily
C, member 18; K09538 DnaJ homolog subfamily C member 18
Length=358
Score = 76.3 bits (186), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 36/81 (44%), Positives = 53/81 (65%), Gaps = 2/81 (2%)
Query 47 IIGAEAARKSKDYYQLLGVRRNASQRAIDKAYRKLAKKLHPD--VAPGKEKEFMEIAKAH 104
++G + +K ++YY++LGV R+AS + KAYRKLA K HPD APG F I A
Sbjct 71 LLGVQRIKKCRNYYEILGVSRDASDEELKKAYRKLALKFHPDKNCAPGATDAFKAIGNAF 130
Query 105 EVLSDPEQRKKYDTFGEEGLA 125
VLS+P++R +YD +G+E +
Sbjct 131 AVLSNPDKRLRYDEYGDEQVT 151
> xla:100037206 dnaja3, tid1; DnaJ (Hsp40) homolog, subfamily
A, member 3; K09504 DnaJ homolog subfamily A member 3
Length=457
Score = 76.3 bits (186), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 37/66 (56%), Positives = 51/66 (77%), Gaps = 3/66 (4%)
Query 58 DYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVA---PGKEKEFMEIAKAHEVLSDPEQRK 114
D+YQ+LGV RNASQ+ I KAY +LAKK HPD P +++F ++A+A+EVLSD +RK
Sbjct 67 DFYQVLGVPRNASQKEIKKAYYQLAKKYHPDTNKEDPQAKEKFSQLAEAYEVLSDEVKRK 126
Query 115 KYDTFG 120
+YDT+G
Sbjct 127 QYDTYG 132
> xla:444375 dnajb12, MGC82876; DnaJ (Hsp40) homolog, subfamily
B, member 12; K09518 DnaJ homolog subfamily B member 12
Length=373
Score = 76.3 bits (186), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 44/112 (39%), Positives = 63/112 (56%), Gaps = 4/112 (3%)
Query 18 NPSSRSSTNSGGPYGAPHSSGGFWSSY--GGIIGAEAARKSKDYYQLLGVRRNASQRAID 75
NP R + P + G SY + + ++ KDYY++LGV R A++ +
Sbjct 68 NPRLRKNAADSTPSANGEAGGETGKSYTPDQLEAVKRIKQCKDYYEILGVTREATEDDLK 127
Query 76 KAYRKLAKKLHPD--VAPGKEKEFMEIAKAHEVLSDPEQRKKYDTFGEEGLA 125
K+YRKLA K HPD APG + F I A+ VLS+ E+RK+YD FGEE ++
Sbjct 128 KSYRKLALKFHPDKNYAPGATEAFKAIGNAYAVLSNAEKRKQYDQFGEEKVS 179
> dre:324005 dnajb12, wu:fc16f06, wu:fi38b10; DnaJ (Hsp40) homolog,
subfamily B, member 12; K09518 DnaJ homolog subfamily
B member 12
Length=371
Score = 75.9 bits (185), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 43/105 (40%), Positives = 60/105 (57%), Gaps = 13/105 (12%)
Query 20 SSRSSTNSGGPYGAPHSSGGFWSSYGGIIGAEAARKSKDYYQLLGVRRNASQRAIDKAYR 79
S S+T S PY + + + ++ KDYY+ LGV + AS+ + KAYR
Sbjct 81 SGHSATESAKPYTSEQ-----------LDAVKRIKRCKDYYETLGVSKEASEEDLKKAYR 129
Query 80 KLAKKLHPDV--APGKEKEFMEIAKAHEVLSDPEQRKKYDTFGEE 122
KLA K HPD APG + F I A+ VLS+PE+R++YD +GEE
Sbjct 130 KLALKFHPDKNHAPGATEAFKAIGNAYAVLSNPEKRRQYDVYGEE 174
> dre:557858 dnajc10, MGC162218, zgc:162218; DnaJ (Hsp40) homolog,
subfamily C, member 10; K09530 DnaJ homolog subfamily C
member 10
Length=791
Score = 75.9 bits (185), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 36/71 (50%), Positives = 51/71 (71%), Gaps = 3/71 (4%)
Query 57 KDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVAPGKEK---EFMEIAKAHEVLSDPEQR 113
+DYY+LLG+ R AS R I +A++KLA +HPD P E +F++I +A+EVL D + R
Sbjct 32 EDYYKLLGISREASTRDIRQAFKKLALTMHPDKNPNDETAHDKFLKINRAYEVLKDEDLR 91
Query 114 KKYDTFGEEGL 124
KKYD +GE+GL
Sbjct 92 KKYDKYGEKGL 102
> hsa:23341 DNAJC16, DKFZp686G1298, DKFZp686N0387, DKFZp781I1547,
KIAA0962; DnaJ (Hsp40) homolog, subfamily C, member 16;
K09536 DnaJ homolog subfamily C member 16
Length=782
Score = 75.9 bits (185), Expect = 5e-14, Method: Composition-based stats.
Identities = 35/68 (51%), Positives = 50/68 (73%), Gaps = 2/68 (2%)
Query 58 DYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVA--PGKEKEFMEIAKAHEVLSDPEQRKK 115
D Y++LGV R ASQ I KAY+KLA++ HPD PG E +F++I+KA+E+LS+ E+R
Sbjct 29 DPYRVLGVSRTASQADIKKAYKKLAREWHPDKNKDPGAEDKFIQISKAYEILSNEEKRSN 88
Query 116 YDTFGEEG 123
YD +G+ G
Sbjct 89 YDQYGDAG 96
> mmu:56323 Dnajb5, 1110058L06Rik, AI462558, Hsc40, Hsp40-3; DnaJ
(Hsp40) homolog, subfamily B, member 5; K09511 DnaJ homolog
subfamily B member 5
Length=348
Score = 75.5 bits (184), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 38/70 (54%), Positives = 52/70 (74%), Gaps = 2/70 (2%)
Query 57 KDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDV--APGKEKEFMEIAKAHEVLSDPEQRK 114
KDYY++LG+ A++ I KAYRK+A K HPD P E++F EIA+A++VLSDP++R
Sbjct 3 KDYYKILGIPSGANEDEIKKAYRKMALKYHPDKNKEPNAEEKFKEIAEAYDVLSDPKKRS 62
Query 115 KYDTFGEEGL 124
YD +GEEGL
Sbjct 63 LYDQYGEEGL 72
> ath:AT3G08910 DNAJ heat shock protein, putative
Length=323
Score = 75.5 bits (184), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 37/72 (51%), Positives = 50/72 (69%), Gaps = 4/72 (5%)
Query 58 DYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVAPGKEKE----FMEIAKAHEVLSDPEQR 113
DYY++L V RNA + KAYRKLA K HPD P +K+ F +I++A++VLSDP++R
Sbjct 4 DYYKVLQVDRNAKDDDLKKAYRKLAMKWHPDKNPNNKKDAEAKFKQISEAYDVLSDPQKR 63
Query 114 KKYDTFGEEGLA 125
YD +GEEGL
Sbjct 64 AIYDQYGEEGLT 75
> xla:734291 dnajb6-a, MGC85133; dnaJ homolog subfamily B member
6-A
Length=250
Score = 75.5 bits (184), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 38/72 (52%), Positives = 51/72 (70%), Gaps = 4/72 (5%)
Query 58 DYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVAPGK----EKEFMEIAKAHEVLSDPEQR 113
+YY++LGV+RNAS I KAYR+LA K HPD P E+ F E+A+A+EVLSD ++R
Sbjct 3 EYYEVLGVQRNASADDIKKAYRRLALKWHPDKNPDNKDEAERRFKEVAEAYEVLSDSKKR 62
Query 114 KKYDTFGEEGLA 125
YD +G+EGL
Sbjct 63 DIYDKYGKEGLT 74
> hsa:4189 DNAJB9, DKFZp564F1862, ERdj4, MDG-1, MDG1, MST049,
MSTP049; DnaJ (Hsp40) homolog, subfamily B, member 9; K09515
DnaJ homolog subfamily B member 9
Length=223
Score = 75.1 bits (183), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 43/100 (43%), Positives = 58/100 (58%), Gaps = 4/100 (4%)
Query 47 IIGAEAARKSKDYYQLLGVRRNASQRAIDKAYRKLAKKLHPD--VAPGKEKEFMEIAKAH 104
++ E SK YY +LGV ++AS+R I KA+ KLA K HPD +P E +F EIA+A+
Sbjct 15 LMITELILASKSYYDILGVPKSASERQIKKAFHKLAMKYHPDKNKSPDAEAKFREIAEAY 74
Query 105 EVLSDPEQRKKYDTFGEEGLAGFPGAGAGDAGFPFSEYVN 144
E LSD +RK+YDT G G G +G F + N
Sbjct 75 ETLSDANRRKEYDTLGHSAFT--SGKGQRGSGSSFEQSFN 112
> ath:AT4G28480 DNAJ heat shock family protein
Length=290
Score = 75.1 bits (183), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 37/73 (50%), Positives = 52/73 (71%), Gaps = 4/73 (5%)
Query 58 DYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVAPGKEKE----FMEIAKAHEVLSDPEQR 113
DYY++L V R+A+ + KAYRKLA K HPD P +K+ F +I++A++VLSDP++R
Sbjct 4 DYYKVLQVDRSANDDDLKKAYRKLAMKWHPDKNPNNKKDAEAKFKQISEAYDVLSDPQKR 63
Query 114 KKYDTFGEEGLAG 126
YD +GEEGL G
Sbjct 64 AVYDQYGEEGLKG 76
> tpv:TP02_0625 chaperone protein DnaJ; K09510 DnaJ homolog subfamily
B member 4
Length=312
Score = 75.1 bits (183), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 38/76 (50%), Positives = 51/76 (67%), Gaps = 6/76 (7%)
Query 57 KDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVAPG------KEKEFMEIAKAHEVLSDP 110
KDYY +LGV+R ++ + KAYRKLA + HPD E+ F +++A++VLSDP
Sbjct 3 KDYYSILGVKRGCNEAELKKAYRKLAMQWHPDKHQDPNSKVKAEEMFKNVSEAYDVLSDP 62
Query 111 EQRKKYDTFGEEGLAG 126
E+RK YD FGEEGL G
Sbjct 63 EKRKIYDQFGEEGLKG 78
> hsa:25822 DNAJB5, Hsc40, KIAA1045; DnaJ (Hsp40) homolog, subfamily
B, member 5; K09511 DnaJ homolog subfamily B member 5
Length=382
Score = 75.1 bits (183), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 39/75 (52%), Positives = 53/75 (70%), Gaps = 2/75 (2%)
Query 52 AARKSKDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDV--APGKEKEFMEIAKAHEVLSD 109
A KDYY++LG+ A++ I KAYRK+A K HPD P E++F EIA+A++VLSD
Sbjct 32 VAVMGKDYYKILGIPSGANEDEIKKAYRKMALKYHPDKNKEPNAEEKFKEIAEAYDVLSD 91
Query 110 PEQRKKYDTFGEEGL 124
P++R YD +GEEGL
Sbjct 92 PKKRGLYDQYGEEGL 106
> ath:AT1G10350 DNAJ heat shock protein, putative
Length=349
Score = 75.1 bits (183), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 35/71 (49%), Positives = 52/71 (73%), Gaps = 4/71 (5%)
Query 58 DYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVAPGKEKE----FMEIAKAHEVLSDPEQR 113
DYY +L V RNA++ + K+YR++A K HPD P +KE F +I++A++VLSDP++R
Sbjct 4 DYYNVLKVNRNANEDDLKKSYRRMAMKWHPDKNPTSKKEAEAKFKQISEAYDVLSDPQRR 63
Query 114 KKYDTFGEEGL 124
+ YD +GEEGL
Sbjct 64 QIYDQYGEEGL 74
> dre:574005 zgc:114162; K09515 DnaJ homolog subfamily B member
9
Length=218
Score = 74.7 bits (182), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 41/95 (43%), Positives = 56/95 (58%), Gaps = 2/95 (2%)
Query 47 IIGAEAARKSKDYYQLLGVRRNASQRAIDKAYRKLAKKLHPD--VAPGKEKEFMEIAKAH 104
++ E KDYY +LGV ++AS+R I KA+ KLA K HPD +P E +F EIA+A+
Sbjct 15 LMITELILARKDYYDILGVPKDASERQIKKAFHKLAMKYHPDKNKSPDAENKFREIAEAY 74
Query 105 EVLSDPEQRKKYDTFGEEGLAGFPGAGAGDAGFPF 139
E LSD ++R++YD G G G AG F
Sbjct 75 ETLSDEKRRREYDRLGHSAFTNDDTNGGGGAGQRF 109
> dre:569466 novel protein similar to vertebrate DnaJ (Hsp40)
homolog, subfamily B, member 5 (DNAJB5); K09511 DnaJ homolog
subfamily B member 5
Length=360
Score = 74.7 bits (182), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 37/70 (52%), Positives = 52/70 (74%), Gaps = 2/70 (2%)
Query 57 KDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDVA--PGKEKEFMEIAKAHEVLSDPEQRK 114
KDYY++LG+ +++ I KAYRK+A K HPD P E++F EIA+A+EVLSDP++R
Sbjct 3 KDYYKILGIPSGSNEDEIKKAYRKMALKFHPDKNKDPNAEEKFKEIAEAYEVLSDPKKRV 62
Query 115 KYDTFGEEGL 124
YD +GE+GL
Sbjct 63 IYDQYGEDGL 72
> cel:F54D5.8 dnj-13; DNaJ domain (prokaryotic heat shock protein)
family member (dnj-13); K09511 DnaJ homolog subfamily B
member 5
Length=331
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 38/70 (54%), Positives = 51/70 (72%), Gaps = 2/70 (2%)
Query 57 KDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDV--APGKEKEFMEIAKAHEVLSDPEQRK 114
KDYY++LG+ + A+ I KAYRK+A K HPD G E +F EIA+A++VLSD +++K
Sbjct 3 KDYYKVLGISKGATDDEIKKAYRKMALKYHPDKNKEAGAENKFKEIAEAYDVLSDDKKKK 62
Query 115 KYDTFGEEGL 124
YD FGEEGL
Sbjct 63 IYDQFGEEGL 72
> mmu:70604 Dnajb14, 5730496F10Rik; DnaJ (Hsp40) homolog, subfamily
B, member 14; K09520 DnaJ homolog subfamily B member 14
Length=379
Score = 74.3 bits (181), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 44/109 (40%), Positives = 61/109 (55%), Gaps = 2/109 (1%)
Query 16 PPNPSSRSSTNSGGPYGAPHSSGGFWSSYGGIIGAEAARKSKDYYQLLGVRRNASQRAID 75
PP S +S + G + GG + + G + K K+YY++LGV ++A +
Sbjct 66 PPGSSDQSKPSCGKDGTSGAGEGGKVYTKDQVEGVLSINKCKNYYEVLGVTKDAGDEDLK 125
Query 76 KAYRKLAKKLHPDV--APGKEKEFMEIAKAHEVLSDPEQRKKYDTFGEE 122
KAYRKLA K HPD APG F +I A+ VLS+PE+RK+YD G E
Sbjct 126 KAYRKLALKFHPDKNHAPGATDAFKKIGNAYAVLSNPEKRKQYDLTGSE 174
> ath:AT1G80030 DNAJ heat shock protein, putative
Length=500
Score = 74.3 bits (181), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 37/80 (46%), Positives = 53/80 (66%), Gaps = 2/80 (2%)
Query 56 SKDYYQLLGVRRNASQRAIDKAYRKLAKKLHPDV--APGKEKEFMEIAKAHEVLSDPEQR 113
S DYY LGV ++A+ + I AYR+LA++ HPDV PG ++F EI+ A+EVLSD ++R
Sbjct 73 SGDYYATLGVSKSANNKEIKAAYRRLARQYHPDVNKEPGATEKFKEISAAYEVLSDEQKR 132
Query 114 KKYDTFGEEGLAGFPGAGAG 133
YD +GE G+ G +G
Sbjct 133 ALYDQYGEAGVKSTVGGASG 152
Lambda K H
0.312 0.134 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2814663556
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40