bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7006_orf1
Length=134
Score E
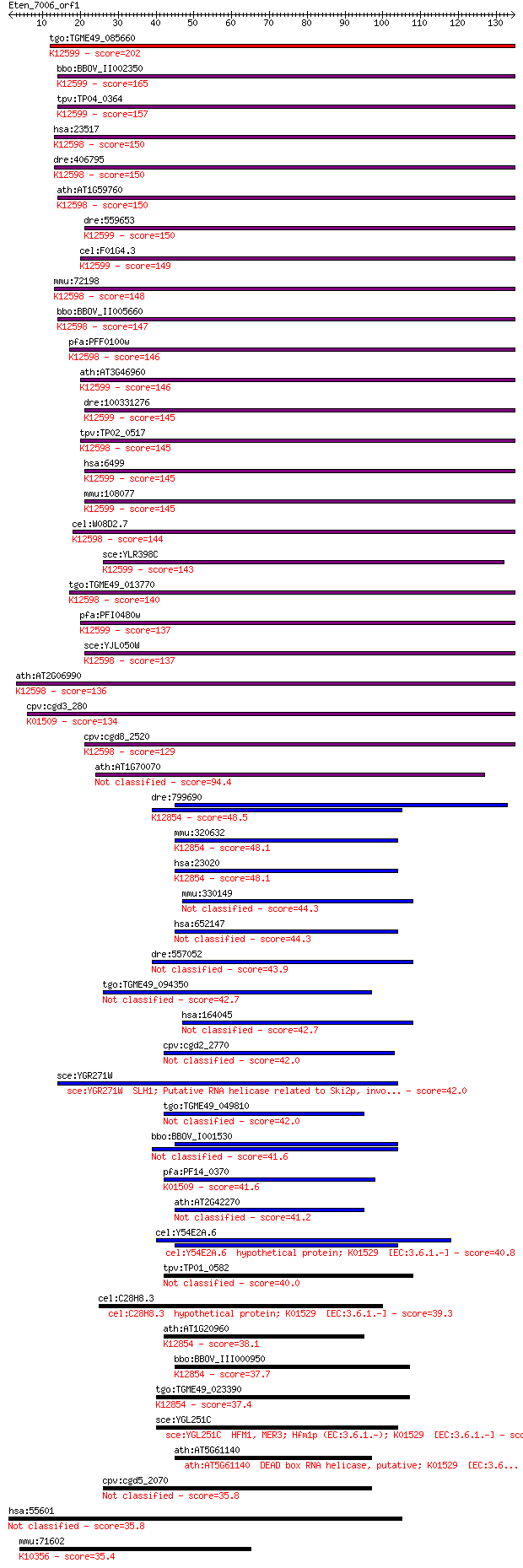
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_085660 DEAD/DEAH box helicase domain-containing pro... 202 3e-52
bbo:BBOV_II002350 18.m06191; helicase with zinc finger motif p... 165 4e-41
tpv:TP04_0364 hypothetical protein; K12599 antiviral helicase ... 157 1e-38
hsa:23517 SKIV2L2, Dob1, KIAA0052, MGC142069, Mtr4, fSAP118; s... 150 7e-37
dre:406795 skiv2l2, wu:fd11a05, zgc:63838; superkiller viralic... 150 7e-37
ath:AT1G59760 ATP-dependent RNA helicase, putative; K12598 ATP... 150 1e-36
dre:559653 skiv2l, fb70b07, wu:fb70b07; superkiller viralicidi... 150 1e-36
cel:F01G4.3 hypothetical protein; K12599 antiviral helicase SK... 149 3e-36
mmu:72198 Skiv2l2, 2610528A15Rik, mKIAA0052; superkiller viral... 148 5e-36
bbo:BBOV_II005660 18.m06470; DSHCT (NUC185) domain containing ... 147 7e-36
pfa:PFF0100w ATP-dependent RNA Helicase, putative (EC:3.6.1.-)... 146 2e-35
ath:AT3G46960 ATP binding / ATP-dependent helicase/ helicase/ ... 146 2e-35
dre:100331276 superkiller viralicidic activity 2-like; K12599 ... 145 2e-35
tpv:TP02_0517 hypothetical protein; K12598 ATP-dependent RNA h... 145 3e-35
hsa:6499 SKIV2L, 170A, DDX13, HLP, SKI2, SKI2W, SKIV2; superki... 145 4e-35
mmu:108077 Skiv2l, 4930534J06Rik, AW214248, Ddx13, SKI, Ski2w;... 145 4e-35
cel:W08D2.7 mtr-4; yeast MTR (mRNA TRansport) homolog family m... 144 8e-35
sce:YLR398C SKI2; Ski complex component and putative RNA helic... 143 2e-34
tgo:TGME49_013770 RNA helicase, putative ; K12598 ATP-dependen... 140 1e-33
pfa:PFI0480w helicase with Zn-finger motif, putative; K12599 a... 137 7e-33
sce:YJL050W MTR4, DOB1; ATP-dependent 3'-5' RNA helicase, invo... 137 8e-33
ath:AT2G06990 HEN2; HEN2 (hua enhancer 2); ATP-dependent helic... 136 2e-32
cpv:cgd3_280 mRNA translation inhibitor SKI2 SFII helicase, DE... 134 8e-32
cpv:cgd8_2520 Mtr4p like SKI family SFII helicase ; K12598 ATP... 129 3e-30
ath:AT1G70070 EMB25; EMB25 (EMBRYO DEFECTIVE 25); ATP-dependen... 94.4 8e-20
dre:799690 fb63a09; wu:fb63a09 (EC:3.6.4.13); K12854 pre-mRNA-... 48.5 5e-06
mmu:320632 Snrnp200, A330064G03Rik, Ascc3l1, BC011390, HELIC2,... 48.1 7e-06
hsa:23020 SNRNP200, ASCC3L1, BRR2, FLJ11521, HELIC2, RP33, U5-... 48.1 8e-06
mmu:330149 Hfm1, A330009G12Rik, Gm1046, Sec63d1; HFM1, ATP-dep... 44.3 1e-04
hsa:652147 u5 small nuclear ribonucleoprotein 200 kDa helicase... 44.3 1e-04
dre:557052 hfm1, si:ch211-83f14.5; HFM1, ATP-dependent DNA hel... 43.9 1e-04
tgo:TGME49_094350 DEAD/DEAH box helicase, putative 42.7 3e-04
hsa:164045 HFM1, FLJ36760, FLJ39011, MER3, MGC163397, RP11-539... 42.7 3e-04
cpv:cgd2_2770 U5 small nuclear ribonucleoprotein 200kDA helica... 42.0 5e-04
sce:YGR271W SLH1; Putative RNA helicase related to Ski2p, invo... 42.0 6e-04
tgo:TGME49_049810 activating signal cointegrator 1 complex sub... 42.0 6e-04
bbo:BBOV_I001530 19.m02171; helicase 41.6 7e-04
pfa:PF14_0370 DEAD/DEAH box helicase, putative; K01509 adenosi... 41.6 7e-04
ath:AT2G42270 U5 small nuclear ribonucleoprotein helicase, put... 41.2 8e-04
cel:Y54E2A.6 hypothetical protein; K01529 [EC:3.6.1.-] 40.8 0.001
tpv:TP01_0582 RNA helicase 40.0 0.002
cel:C28H8.3 hypothetical protein; K01529 [EC:3.6.1.-] 39.3 0.004
ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding / A... 38.1 0.007
bbo:BBOV_III000950 17.m07111; sec63 domain containing protein ... 37.7 0.011
tgo:TGME49_023390 sec63 domain-containing DEAD/DEAH box helica... 37.4 0.014
sce:YGL251C HFM1, MER3; Hfm1p (EC:3.6.1.-); K01529 [EC:3.6.1.-] 36.6 0.024
ath:AT5G61140 DEAD box RNA helicase, putative; K01529 [EC:3.6... 36.2 0.027
cpv:cgd5_2070 SPAC694.02. SKI family SFII helicase 35.8 0.034
hsa:55601 DDX60, FLJ10787, FLJ20035; DEAD (Asp-Glu-Ala-Asp) bo... 35.8 0.039
mmu:71602 Myo1e, 2310020N23Rik, 9130023P14Rik, AA407778, myosi... 35.4 0.045
> tgo:TGME49_085660 DEAD/DEAH box helicase domain-containing protein
; K12599 antiviral helicase SKI2 [EC:3.6.4.-]
Length=1329
Score = 202 bits (513), Expect = 3e-52, Method: Compositional matrix adjust.
Identities = 96/123 (78%), Positives = 109/123 (88%), Gaps = 0/123 (0%)
Query 12 DLSPEEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRR 71
DLSP + PLA+EF FPLDDFQKRAIL LEK Q VFVAAHTSAGKTVVAEYAIALA++R R
Sbjct 442 DLSPLQVPLALEFPFPLDDFQKRAILHLEKYQTVFVAAHTSAGKTVVAEYAIALAVRRNR 501
Query 72 RCIFTSPLKALSNQKYRDFKQKFVSVGLITGDICINPGAACLIVTTEILRSLLFRGDEII 131
RCI+TSPLKALSNQKYR+F+ KF SVG++TGD+CINP A CLIVTTEILRSLL+ GD +I
Sbjct 502 RCIYTSPLKALSNQKYREFRLKFPSVGIVTGDVCINPDANCLIVTTEILRSLLYLGDALI 561
Query 132 SQL 134
Q+
Sbjct 562 GQV 564
> bbo:BBOV_II002350 18.m06191; helicase with zinc finger motif
protein; K12599 antiviral helicase SKI2 [EC:3.6.4.-]
Length=1113
Score = 165 bits (417), Expect = 4e-41, Method: Composition-based stats.
Identities = 81/123 (65%), Positives = 100/123 (81%), Gaps = 2/123 (1%)
Query 14 SPEEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRRC 73
+PE E L IE+ F LDDFQKRAI L K + VFVAAHTS+GKTVVAEYAIALAL R ++
Sbjct 195 TPELEDLVIEYPFELDDFQKRAIYHLHKMKHVFVAAHTSSGKTVVAEYAIALALSRGKKA 254
Query 74 IFTSPLKALSNQKYRDFKQKF--VSVGLITGDICINPGAACLIVTTEILRSLLFRGDEII 131
++TSP+KALSNQK+R+F +++ +VG+ITGD+ NP A CLIVTTEILR+LL+RGD II
Sbjct 255 VYTSPIKALSNQKFREFTKRYGNETVGIITGDVSCNPNAPCLIVTTEILRNLLYRGDPII 314
Query 132 SQL 134
QL
Sbjct 315 GQL 317
> tpv:TP04_0364 hypothetical protein; K12599 antiviral helicase
SKI2 [EC:3.6.4.-]
Length=1069
Score = 157 bits (396), Expect = 1e-38, Method: Composition-based stats.
Identities = 77/123 (62%), Positives = 99/123 (80%), Gaps = 2/123 (1%)
Query 14 SPEEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRRC 73
+PE L ++ F LDDFQK++I L + VFV+AHTSAGKTVVAEY+IALA+ R ++
Sbjct 169 APELTDLIAQYPFELDDFQKKSIHHLINGKHVFVSAHTSAGKTVVAEYSIALAISRGQKA 228
Query 74 IFTSPLKALSNQKYRDFKQKF--VSVGLITGDICINPGAACLIVTTEILRSLLFRGDEII 131
I+TSP+KALSNQKYR+FK KF +VG+ITGD+ NPGA+CLIVTTEILR+LL+RGD +I
Sbjct 229 IYTSPIKALSNQKYREFKVKFGNENVGIITGDVLCNPGASCLIVTTEILRNLLYRGDSVI 288
Query 132 SQL 134
Q+
Sbjct 289 GQI 291
> hsa:23517 SKIV2L2, Dob1, KIAA0052, MGC142069, Mtr4, fSAP118;
superkiller viralicidic activity 2-like 2 (S. cerevisiae) (EC:3.6.4.13);
K12598 ATP-dependent RNA helicase DOB1 [EC:3.6.4.13]
Length=1042
Score = 150 bits (380), Expect = 7e-37, Method: Compositional matrix adjust.
Identities = 72/122 (59%), Positives = 95/122 (77%), Gaps = 0/122 (0%)
Query 13 LSPEEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRR 72
L P A E+ F LD FQ+ AI ++ NQ V V+AHTSAGKTV AEYAIALAL+ ++R
Sbjct 125 LKPRVGKAAKEYPFILDAFQREAIQCVDNNQSVLVSAHTSAGKTVCAEYAIALALREKQR 184
Query 73 CIFTSPLKALSNQKYRDFKQKFVSVGLITGDICINPGAACLIVTTEILRSLLFRGDEIIS 132
IFTSP+KALSNQKYR+ ++F VGL+TGD+ INP A+CL++TTEILRS+L+RG E++
Sbjct 185 VIFTSPIKALSNQKYREMYEEFQDVGLMTGDVTINPTASCLVMTTEILRSMLYRGSEVMR 244
Query 133 QL 134
++
Sbjct 245 EV 246
> dre:406795 skiv2l2, wu:fd11a05, zgc:63838; superkiller viralicidic
activity 2-like 2 (EC:3.6.4.13); K12598 ATP-dependent
RNA helicase DOB1 [EC:3.6.4.13]
Length=1034
Score = 150 bits (380), Expect = 7e-37, Method: Composition-based stats.
Identities = 73/122 (59%), Positives = 96/122 (78%), Gaps = 0/122 (0%)
Query 13 LSPEEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRR 72
L P A E+ F LD FQ+ AIL ++ NQ V V+AHTSAGKTV AEYAIALAL+ ++R
Sbjct 113 LKPRVGKAAKEYPFILDPFQREAILCIDNNQSVLVSAHTSAGKTVCAEYAIALALREKQR 172
Query 73 CIFTSPLKALSNQKYRDFKQKFVSVGLITGDICINPGAACLIVTTEILRSLLFRGDEIIS 132
IFTSP+KALSNQKYR+ ++F VGL+TGD+ INP A+CL++TTEILRS+L+RG E++
Sbjct 173 VIFTSPIKALSNQKYREMYEEFQDVGLMTGDVTINPTASCLVMTTEILRSMLYRGSEVMR 232
Query 133 QL 134
++
Sbjct 233 EV 234
> ath:AT1G59760 ATP-dependent RNA helicase, putative; K12598 ATP-dependent
RNA helicase DOB1 [EC:3.6.4.13]
Length=988
Score = 150 bits (379), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 69/121 (57%), Positives = 94/121 (77%), Gaps = 0/121 (0%)
Query 14 SPEEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRRC 73
S +P A +F F LD FQ AI L+ + V V+AHTSAGKTVVA YAIA++L+ +R
Sbjct 54 SVHNKPPAKDFPFTLDSFQSEAIKCLDNGESVMVSAHTSAGKTVVASYAIAMSLKENQRV 113
Query 74 IFTSPLKALSNQKYRDFKQKFVSVGLITGDICINPGAACLIVTTEILRSLLFRGDEIISQ 133
I+TSP+KALSNQKYRDFK++F VGL+TGD+ I+P A+CL++TTEILRS+ ++G EI+ +
Sbjct 114 IYTSPIKALSNQKYRDFKEEFSDVGLMTGDVTIDPNASCLVMTTEILRSMQYKGSEIMRE 173
Query 134 L 134
+
Sbjct 174 V 174
> dre:559653 skiv2l, fb70b07, wu:fb70b07; superkiller viralicidic
activity 2 (S. cerevisiae homolog)-like; K12599 antiviral
helicase SKI2 [EC:3.6.4.-]
Length=1230
Score = 150 bits (378), Expect = 1e-36, Method: Composition-based stats.
Identities = 74/114 (64%), Positives = 91/114 (79%), Gaps = 0/114 (0%)
Query 21 AIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRRCIFTSPLK 80
A ++ F LD FQK+AILRLE + VFVAAHTSAGKTVVAEYAIAL+ + R I+TSP+K
Sbjct 312 AFKYPFELDVFQKQAILRLEAHDSVFVAAHTSAGKTVVAEYAIALSQKHMTRTIYTSPIK 371
Query 81 ALSNQKYRDFKQKFVSVGLITGDICINPGAACLIVTTEILRSLLFRGDEIISQL 134
ALSNQK+RDFK F VGL+TGD+ +NP +CLI+TTEILRS+L+ G E+I L
Sbjct 372 ALSNQKFRDFKNTFGDVGLLTGDVQLNPEGSCLIMTTEILRSMLYNGSEVIRDL 425
> cel:F01G4.3 hypothetical protein; K12599 antiviral helicase
SKI2 [EC:3.6.4.-]
Length=1266
Score = 149 bits (376), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 70/115 (60%), Positives = 92/115 (80%), Gaps = 0/115 (0%)
Query 20 LAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRRCIFTSPL 79
+A ++ F LD FQ+ ++L +E+ + +FVAAHTSAGKTVVAEYAIAL + R ++TSP+
Sbjct 285 MARKYPFSLDPFQQSSVLCMERGESLFVAAHTSAGKTVVAEYAIALCQAHKTRAVYTSPI 344
Query 80 KALSNQKYRDFKQKFVSVGLITGDICINPGAACLIVTTEILRSLLFRGDEIISQL 134
KALSNQK+RDFKQ F VGL+TGDI ++P AACLI+TTEILRS+L+ G E+I L
Sbjct 345 KALSNQKFRDFKQIFGDVGLVTGDIQLHPEAACLIMTTEILRSMLYNGSEVIRDL 399
> mmu:72198 Skiv2l2, 2610528A15Rik, mKIAA0052; superkiller viralicidic
activity 2-like 2 (S. cerevisiae) (EC:3.6.4.13); K12598
ATP-dependent RNA helicase DOB1 [EC:3.6.4.13]
Length=1040
Score = 148 bits (373), Expect = 5e-36, Method: Composition-based stats.
Identities = 72/122 (59%), Positives = 95/122 (77%), Gaps = 0/122 (0%)
Query 13 LSPEEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRR 72
L P A E+ F LD FQ+ AI ++ NQ V V+AHTSAGKTV AEYAIALAL+ ++R
Sbjct 123 LKPRVGKAAKEYPFILDAFQREAIQCVDNNQSVLVSAHTSAGKTVCAEYAIALALREKQR 182
Query 73 CIFTSPLKALSNQKYRDFKQKFVSVGLITGDICINPGAACLIVTTEILRSLLFRGDEIIS 132
IFTSP+KALSNQKYR+ ++F VGL+TGD+ INP A+CL++TTEILRS+L+RG E++
Sbjct 183 VIFTSPIKALSNQKYREMYEEFQDVGLMTGDVTINPTASCLVMTTEILRSMLYRGSEVMR 242
Query 133 QL 134
++
Sbjct 243 EV 244
> bbo:BBOV_II005660 18.m06470; DSHCT (NUC185) domain containing
DEAD/DEAH box helicase family protein (EC:3.6.1.3); K12598
ATP-dependent RNA helicase DOB1 [EC:3.6.4.13]
Length=986
Score = 147 bits (372), Expect = 7e-36, Method: Composition-based stats.
Identities = 73/123 (59%), Positives = 96/123 (78%), Gaps = 2/123 (1%)
Query 14 SPEEEP--LAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRR 71
+P EP A + F LDDFQKR+I LEK + V V AHTSAGKTVVAEYAIA+ L+ +R
Sbjct 68 TPPYEPGEPARSYPFTLDDFQKRSIECLEKGESVLVCAHTSAGKTVVAEYAIAMGLRDKR 127
Query 72 RCIFTSPLKALSNQKYRDFKQKFVSVGLITGDICINPGAACLIVTTEILRSLLFRGDEII 131
R I+TSP+KALSNQKYR+ +FV VGL+TGD+ +NP A+ +++TTEILRS+L+RG EI+
Sbjct 128 RIIYTSPIKALSNQKYRNLCDEFVDVGLMTGDVTLNPDASVMVMTTEILRSMLYRGSEIV 187
Query 132 SQL 134
++
Sbjct 188 QEM 190
> pfa:PFF0100w ATP-dependent RNA Helicase, putative (EC:3.6.1.-);
K12598 ATP-dependent RNA helicase DOB1 [EC:3.6.4.13]
Length=1350
Score = 146 bits (368), Expect = 2e-35, Method: Composition-based stats.
Identities = 71/120 (59%), Positives = 95/120 (79%), Gaps = 2/120 (1%)
Query 17 EEPL--AIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRRCI 74
+EPL A + F LD FQK++I LE+N+ V V+AHTSAGKTV+AEYAIAL L+ ++R I
Sbjct 242 KEPLIPARTYKFELDTFQKKSIECLERNESVLVSAHTSAGKTVIAEYAIALGLRDKQRVI 301
Query 75 FTSPLKALSNQKYRDFKQKFVSVGLITGDICINPGAACLIVTTEILRSLLFRGDEIISQL 134
+TSP+KALSNQKYRD ++F VGLITGDI INP A+ +++TTEILRS+L+RG + ++
Sbjct 302 YTSPIKALSNQKYRDLGEEFKDVGLITGDISINPEASIIVMTTEILRSMLYRGSSLTKEV 361
> ath:AT3G46960 ATP binding / ATP-dependent helicase/ helicase/
hydrolase, acting on acid anhydrides, in phosphorus-containing
anhydrides / nucleic acid binding; K12599 antiviral helicase
SKI2 [EC:3.6.4.-]
Length=1347
Score = 146 bits (368), Expect = 2e-35, Method: Composition-based stats.
Identities = 74/115 (64%), Positives = 91/115 (79%), Gaps = 1/115 (0%)
Query 20 LAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRRCIFTSPL 79
+AIEF F LD+FQK AI LEK + VFVAAHTSAGKTVVAEYA ALA + R ++T+P+
Sbjct 353 MAIEFPFELDNFQKEAICCLEKGESVFVAAHTSAGKTVVAEYAFALATKHCTRAVYTAPI 412
Query 80 KALSNQKYRDFKQKFVSVGLITGDICINPGAACLIVTTEILRSLLFRGDEIISQL 134
K +SNQKYRDF KF VGL+TGD+ I P A+CLI+TTEILRS+L+RG +II +
Sbjct 413 KTISNQKYRDFCGKF-DVGLLTGDVSIRPEASCLIMTTEILRSMLYRGADIIRDI 466
> dre:100331276 superkiller viralicidic activity 2-like; K12599
antiviral helicase SKI2 [EC:3.6.4.-]
Length=1235
Score = 145 bits (367), Expect = 2e-35, Method: Composition-based stats.
Identities = 71/114 (62%), Positives = 91/114 (79%), Gaps = 0/114 (0%)
Query 21 AIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRRCIFTSPLK 80
A ++ F D FQK+AIL LE++ VFVAAHTSAGKTVVAEYAIALA + R I+TSP+K
Sbjct 304 AFQWAFEPDVFQKQAILHLERHDSVFVAAHTSAGKTVVAEYAIALAQKHMTRTIYTSPIK 363
Query 81 ALSNQKYRDFKQKFVSVGLITGDICINPGAACLIVTTEILRSLLFRGDEIISQL 134
ALSNQK+RDF+ F VGL+TGD+ ++P A+CLI+TTEILRS+L+ G ++I L
Sbjct 364 ALSNQKFRDFRNTFGDVGLLTGDVQLHPEASCLIMTTEILRSMLYSGSDVIRDL 417
> tpv:TP02_0517 hypothetical protein; K12598 ATP-dependent RNA
helicase DOB1 [EC:3.6.4.13]
Length=1012
Score = 145 bits (367), Expect = 3e-35, Method: Composition-based stats.
Identities = 67/115 (58%), Positives = 92/115 (80%), Gaps = 0/115 (0%)
Query 20 LAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRRCIFTSPL 79
A ++ F LD+FQKR+I LE+++ V V AHTSAGKTVVAEYAIA+ L+ R I+TSP+
Sbjct 100 FAKKYPFTLDEFQKRSIESLERDESVLVCAHTSAGKTVVAEYAIAMGLRDGHRIIYTSPI 159
Query 80 KALSNQKYRDFKQKFVSVGLITGDICINPGAACLIVTTEILRSLLFRGDEIISQL 134
KALSNQKYR+ +FV VGL+TGD+ +NP A+ +++TTEILRS+L+RG EI+ ++
Sbjct 160 KALSNQKYRNLSDEFVDVGLMTGDVTLNPNASVMVMTTEILRSMLYRGSEIVQEM 214
> hsa:6499 SKIV2L, 170A, DDX13, HLP, SKI2, SKI2W, SKIV2; superkiller
viralicidic activity 2-like (S. cerevisiae); K12599 antiviral
helicase SKI2 [EC:3.6.4.-]
Length=1246
Score = 145 bits (365), Expect = 4e-35, Method: Composition-based stats.
Identities = 71/114 (62%), Positives = 91/114 (79%), Gaps = 0/114 (0%)
Query 21 AIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRRCIFTSPLK 80
A ++ F D FQK+AIL LE++ VFVAAHTSAGKTVVAEYAIALA + R I+TSP+K
Sbjct 304 AFQWAFEPDVFQKQAILHLERHDSVFVAAHTSAGKTVVAEYAIALAQKHMTRTIYTSPIK 363
Query 81 ALSNQKYRDFKQKFVSVGLITGDICINPGAACLIVTTEILRSLLFRGDEIISQL 134
ALSNQK+RDF+ F VGL+TGD+ ++P A+CLI+TTEILRS+L+ G ++I L
Sbjct 364 ALSNQKFRDFRNTFGDVGLLTGDVQLHPEASCLIMTTEILRSMLYSGSDVIRDL 417
> mmu:108077 Skiv2l, 4930534J06Rik, AW214248, Ddx13, SKI, Ski2w;
superkiller viralicidic activity 2-like (S. cerevisiae);
K12599 antiviral helicase SKI2 [EC:3.6.4.-]
Length=1244
Score = 145 bits (365), Expect = 4e-35, Method: Composition-based stats.
Identities = 71/114 (62%), Positives = 91/114 (79%), Gaps = 0/114 (0%)
Query 21 AIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRRCIFTSPLK 80
A ++ F D FQK+AIL LE++ VFVAAHTSAGKTVVAEYAIALA + R I+TSP+K
Sbjct 301 AFQWAFEPDVFQKQAILHLEQHDSVFVAAHTSAGKTVVAEYAIALAQKHMTRTIYTSPIK 360
Query 81 ALSNQKYRDFKQKFVSVGLITGDICINPGAACLIVTTEILRSLLFRGDEIISQL 134
ALSNQK+RDF+ F VGL+TGD+ ++P A+CLI+TTEILRS+L+ G ++I L
Sbjct 361 ALSNQKFRDFRNTFGDVGLLTGDVQLHPEASCLIMTTEILRSMLYSGSDVIRDL 414
> cel:W08D2.7 mtr-4; yeast MTR (mRNA TRansport) homolog family
member (mtr-4); K12598 ATP-dependent RNA helicase DOB1 [EC:3.6.4.13]
Length=1026
Score = 144 bits (363), Expect = 8e-35, Method: Composition-based stats.
Identities = 70/117 (59%), Positives = 95/117 (81%), Gaps = 1/117 (0%)
Query 18 EPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRRCIFTS 77
EP A + F LD FQK+AIL ++ NQ V V+AHTSAGKTVVA YAIA L+ ++R I+TS
Sbjct 117 EP-AKYYPFQLDAFQKQAILCIDNNQSVLVSAHTSAGKTVVATYAIAKCLREKQRVIYTS 175
Query 78 PLKALSNQKYRDFKQKFVSVGLITGDICINPGAACLIVTTEILRSLLFRGDEIISQL 134
P+KALSNQKYR+ +++F VGL+TGD+ +NP A+CL++TTEILRS+L+RG EI+ ++
Sbjct 176 PIKALSNQKYRELEEEFKDVGLMTGDVTLNPDASCLVMTTEILRSMLYRGSEIMKEV 232
> sce:YLR398C SKI2; Ski complex component and putative RNA helicase,
mediates 3'-5' RNA degradation by the cytoplasmic exosome;
null mutants have superkiller phenotype of increased viral
dsRNAs and are synthetic lethal with mutations in 5'-3'
mRNA decay (EC:3.6.1.-); K12599 antiviral helicase SKI2 [EC:3.6.4.-]
Length=1287
Score = 143 bits (360), Expect = 2e-34, Method: Composition-based stats.
Identities = 71/108 (65%), Positives = 88/108 (81%), Gaps = 2/108 (1%)
Query 26 FPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRRCIFTSPLKALSNQ 85
F LD FQK A+ LE+ VFVAAHTSAGKTVVAEYAIA+A + + I+TSP+KALSNQ
Sbjct 328 FELDTFQKEAVYHLEQGDSVFVAAHTSAGKTVVAEYAIAMAHRNMTKTIYTSPIKALSNQ 387
Query 86 KYRDFKQKF--VSVGLITGDICINPGAACLIVTTEILRSLLFRGDEII 131
K+RDFK+ F V++GLITGD+ INP A CLI+TTEILRS+L+RG ++I
Sbjct 388 KFRDFKETFDDVNIGLITGDVQINPDANCLIMTTEILRSMLYRGADLI 435
> tgo:TGME49_013770 RNA helicase, putative ; K12598 ATP-dependent
RNA helicase DOB1 [EC:3.6.4.13]
Length=1206
Score = 140 bits (352), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 65/120 (54%), Positives = 97/120 (80%), Gaps = 3/120 (2%)
Query 17 EEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRRCIFT 76
+EP A ++ F LD FQ+R++L LE + V VAAHTSAGKTVVAEYAIA++++ ++R ++T
Sbjct 225 KEP-ARQYKFRLDAFQQRSVLCLEAGESVLVAAHTSAGKTVVAEYAIAMSIRDKQRVVYT 283
Query 77 SPLKALSNQKYRDFKQKFV--SVGLITGDICINPGAACLIVTTEILRSLLFRGDEIISQL 134
SP+KALSNQKYRD + F +VGL+TGD+ ++P A+ +++TTEILRS+L+RG ++++L
Sbjct 284 SPIKALSNQKYRDLSESFGAENVGLMTGDVTLHPNASIMVMTTEILRSMLYRGSHLVNEL 343
> pfa:PFI0480w helicase with Zn-finger motif, putative; K12599
antiviral helicase SKI2 [EC:3.6.4.-]
Length=1373
Score = 137 bits (346), Expect = 7e-33, Method: Composition-based stats.
Identities = 65/115 (56%), Positives = 90/115 (78%), Gaps = 0/115 (0%)
Query 20 LAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRRCIFTSPL 79
L + + F LDDFQKR+I L + VFVAAHTSAGKT++AE+AIAL+++ +++ I+TSP+
Sbjct 294 LLLNYDFELDDFQKRSIKHLNNFKHVFVAAHTSAGKTLIAEHAIALSIKLQKKAIYTSPI 353
Query 80 KALSNQKYRDFKQKFVSVGLITGDICINPGAACLIVTTEILRSLLFRGDEIISQL 134
KALSNQKY +FK F VG+ITGD+ +N A C+I+TTEILR+LL+ D II+ +
Sbjct 354 KALSNQKYYEFKNIFKDVGIITGDVKMNVNANCIIMTTEILRNLLYLNDNIINNI 408
> sce:YJL050W MTR4, DOB1; ATP-dependent 3'-5' RNA helicase, involved
in nuclear RNA processing and degredation both as a component
of the TRAMP complex and in TRAMP independent processes;
member of the Dead-box family of helicases (EC:3.6.1.-);
K12598 ATP-dependent RNA helicase DOB1 [EC:3.6.4.13]
Length=1073
Score = 137 bits (346), Expect = 8e-33, Method: Composition-based stats.
Identities = 66/114 (57%), Positives = 89/114 (78%), Gaps = 0/114 (0%)
Query 21 AIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRRCIFTSPLK 80
A + F LD FQ AI +++ + V V+AHTSAGKTVVAEYAIA +L+ ++R I+TSP+K
Sbjct 143 ARTYPFTLDPFQDTAISCIDRGESVLVSAHTSAGKTVVAEYAIAQSLKNKQRVIYTSPIK 202
Query 81 ALSNQKYRDFKQKFVSVGLITGDICINPGAACLIVTTEILRSLLFRGDEIISQL 134
ALSNQKYR+ +F VGL+TGDI INP A CL++TTEILRS+L+RG E++ ++
Sbjct 203 ALSNQKYRELLAEFGDVGLMTGDITINPDAGCLVMTTEILRSMLYRGSEVMREV 256
> ath:AT2G06990 HEN2; HEN2 (hua enhancer 2); ATP-dependent helicase/
RNA helicase; K12598 ATP-dependent RNA helicase DOB1
[EC:3.6.4.13]
Length=995
Score = 136 bits (343), Expect = 2e-32, Method: Composition-based stats.
Identities = 65/132 (49%), Positives = 96/132 (72%), Gaps = 0/132 (0%)
Query 3 EEEEAAATQDLSPEEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYA 62
+EE T D +A + F LD FQ ++ LE+ + + V+AHTSAGKT VAEYA
Sbjct 57 KEETIHGTLDNPVFNGDMAKTYPFKLDPFQSVSVACLERKESILVSAHTSAGKTAVAEYA 116
Query 63 IALALQRRRRCIFTSPLKALSNQKYRDFKQKFVSVGLITGDICINPGAACLIVTTEILRS 122
IA+A + ++R I+TSPLKALSNQKYR+ + +F VGL+TGD+ ++P A+CL++TTEILR+
Sbjct 117 IAMAFRDKQRVIYTSPLKALSNQKYRELQHEFKDVGLMTGDVTLSPNASCLVMTTEILRA 176
Query 123 LLFRGDEIISQL 134
+L+RG E++ ++
Sbjct 177 MLYRGSEVLKEV 188
> cpv:cgd3_280 mRNA translation inhibitor SKI2 SFII helicase,
DEXDc+HELICc ; K01509 adenosinetriphosphatase [EC:3.6.1.3]
Length=1439
Score = 134 bits (337), Expect = 8e-32, Method: Composition-based stats.
Identities = 69/143 (48%), Positives = 94/143 (65%), Gaps = 14/143 (9%)
Query 6 EAAATQDLSPEE--EPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAI 63
E +DL E E + +++ + LDDFQKRA++ + V VAAHTSAGKT VAEYAI
Sbjct 104 EPCKKEDLDDSEDLENVVLKYPYELDDFQKRAVINIHNGDHVLVAAHTSAGKTAVAEYAI 163
Query 64 ALALQRRRRCIFTSPLKALSNQKYRDFKQKFVS------------VGLITGDICINPGAA 111
LA + R+ I+TSP+KALS+QKYR+F +F +G+ITGD+ INP A
Sbjct 164 ELANKNGRKAIYTSPIKALSSQKYREFLNRFREYPAHSSFTQRNRIGIITGDVSINPDAQ 223
Query 112 CLIVTTEILRSLLFRGDEIISQL 134
C+I+TTEILR++L+R D I Q+
Sbjct 224 CVIMTTEILRTMLYRNDPCIEQI 246
> cpv:cgd8_2520 Mtr4p like SKI family SFII helicase ; K12598 ATP-dependent
RNA helicase DOB1 [EC:3.6.4.13]
Length=1280
Score = 129 bits (324), Expect = 3e-30, Method: Composition-based stats.
Identities = 62/116 (53%), Positives = 88/116 (75%), Gaps = 2/116 (1%)
Query 21 AIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRRCIFTSPLK 80
A E+ F LD FQ +I LE + V ++AHTSAGKT VAEYAIA+AL+ +R ++TSP+K
Sbjct 93 AKEYSFKLDTFQAVSIGCLEIGESVLISAHTSAGKTCVAEYAIAMALKSNQRVVYTSPIK 152
Query 81 ALSNQKYRDFKQKF--VSVGLITGDICINPGAACLIVTTEILRSLLFRGDEIISQL 134
ALSNQKYRD + F +VGL+TGD+ +NP + +++TTEILRS+L+RG E++ ++
Sbjct 153 ALSNQKYRDLRTTFGDNNVGLLTGDVTVNPLGSIMVMTTEILRSMLYRGSELVREI 208
> ath:AT1G70070 EMB25; EMB25 (EMBRYO DEFECTIVE 25); ATP-dependent
helicase/ RNA helicase
Length=1171
Score = 94.4 bits (233), Expect = 8e-20, Method: Composition-based stats.
Identities = 49/105 (46%), Positives = 73/105 (69%), Gaps = 2/105 (1%)
Query 24 FGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRRCIFTSPLKALS 83
+ F +D FQ+ AI + V V+A TS+GKT++AE A + + RR +T+PLKALS
Sbjct 151 YDFRIDKFQRLAIEAFLRGSSVVVSAPTSSGKTLIAEAAAVSTVAKGRRLFYTTPLKALS 210
Query 84 NQKYRDFKQKF--VSVGLITGDICINPGAACLIVTTEILRSLLFR 126
NQK+R+F++ F +VGL+TGD IN A +I+TTEILR++L++
Sbjct 211 NQKFREFRETFGDDNVGLLTGDSAINKDAQIVIMTTEILRNMLYQ 255
> dre:799690 fb63a09; wu:fb63a09 (EC:3.6.4.13); K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2134
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 32/90 (35%), Positives = 46/90 (51%), Gaps = 10/90 (11%)
Query 45 VFVAAHTSAGKTVVAEYAIALAL--QRRRRCIFTSPLKALSNQKYRDFKQKFVSVGLITG 102
VFV A T +GKT+ AE+AI L RC++ +P++AL+ Q + D+ QKF
Sbjct 1342 VFVGAPTGSGKTICAEFAILRMLLHNAEGRCVYITPMEALAEQVFLDWHQKFQE------ 1395
Query 103 DICINPGAACLIVTTEILRSLLFRGDEIIS 132
+N L T LL +GD I+S
Sbjct 1396 --NLNKKVVLLTGETSTDLKLLGKGDIIVS 1423
Score = 29.3 bits (64), Expect = 3.6, Method: Composition-based stats.
Identities = 18/77 (23%), Positives = 37/77 (48%), Gaps = 11/77 (14%)
Query 39 LEKNQCVFVAAHTSAGKTVVAEYAIALALQRR-----------RRCIFTSPLKALSNQKY 87
+E ++ + V A T AGKT VA A+ + + + I+ +P+++L +
Sbjct 490 METDENLLVCAPTGAGKTNVALMAMLREIGKHINMDGTINVADFKIIYIAPMRSLVQEMV 549
Query 88 RDFKQKFVSVGLITGDI 104
F ++ S G+I ++
Sbjct 550 GSFGKRLASYGIIVSEL 566
> mmu:320632 Snrnp200, A330064G03Rik, Ascc3l1, BC011390, HELIC2,
KIAA0788, U5-200-KD, U5-200KD; small nuclear ribonucleoprotein
200 (U5) (EC:3.6.4.13); K12854 pre-mRNA-splicing helicase
BRR2 [EC:3.6.4.13]
Length=2136
Score = 48.1 bits (113), Expect = 7e-06, Method: Composition-based stats.
Identities = 27/66 (40%), Positives = 41/66 (62%), Gaps = 7/66 (10%)
Query 45 VFVAAHTSAGKTVVAEYAIALALQRRR--RCIFTSPLKALSNQKYRDFKQKFV-----SV 97
VFV A T +GKT+ AE+AI L + RC++ +P++AL+ Q Y D+ +KF V
Sbjct 1346 VFVGAPTGSGKTICAEFAILRMLLQNSEGRCVYITPMEALAEQVYMDWYEKFQDRLNKKV 1405
Query 98 GLITGD 103
L+TG+
Sbjct 1406 VLLTGE 1411
> hsa:23020 SNRNP200, ASCC3L1, BRR2, FLJ11521, HELIC2, RP33, U5-200KD;
small nuclear ribonucleoprotein 200kDa (U5) (EC:3.6.4.13);
K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2136
Score = 48.1 bits (113), Expect = 8e-06, Method: Composition-based stats.
Identities = 27/66 (40%), Positives = 41/66 (62%), Gaps = 7/66 (10%)
Query 45 VFVAAHTSAGKTVVAEYAIALALQRRR--RCIFTSPLKALSNQKYRDFKQKFV-----SV 97
VFV A T +GKT+ AE+AI L + RC++ +P++AL+ Q Y D+ +KF V
Sbjct 1346 VFVGAPTGSGKTICAEFAILRMLLQSSEGRCVYITPMEALAEQVYMDWYEKFQDRLNKKV 1405
Query 98 GLITGD 103
L+TG+
Sbjct 1406 VLLTGE 1411
> mmu:330149 Hfm1, A330009G12Rik, Gm1046, Sec63d1; HFM1, ATP-dependent
DNA helicase homolog (S. cerevisiae)
Length=1434
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 26/71 (36%), Positives = 41/71 (57%), Gaps = 10/71 (14%)
Query 47 VAAHTSAGKTVVAEYAIALALQR------RRRCIFTSPLKALSNQKYRDFKQKFVSVGL- 99
+ A T +GKTVV E AI L + ++ +P+KAL +Q++ D+K+KF VGL
Sbjct 300 ICAPTGSGKTVVFELAITRLLMEVPLPWLNMKIVYMAPIKALCSQRFDDWKEKFGPVGLN 359
Query 100 ---ITGDICIN 107
+TGD ++
Sbjct 360 CKELTGDTVMD 370
> hsa:652147 u5 small nuclear ribonucleoprotein 200 kDa helicase-like
Length=1700
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 25/66 (37%), Positives = 38/66 (57%), Gaps = 7/66 (10%)
Query 45 VFVAAHTSAGKTVVAEYAIALALQRRR--RCIFTSPLKALSNQKYRDFKQKFV-----SV 97
VFV A T +GKT+ AE+AI L + RC++ +P++ Q Y D+ +KF V
Sbjct 910 VFVGAPTGSGKTICAEFAILRMLLQNSEGRCVYITPMRLWQEQVYMDWYEKFQDRLNKKV 969
Query 98 GLITGD 103
L+TG+
Sbjct 970 VLLTGE 975
> dre:557052 hfm1, si:ch211-83f14.5; HFM1, ATP-dependent DNA helicase
homolog (S. cerevisiae)
Length=1228
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 29/79 (36%), Positives = 43/79 (54%), Gaps = 10/79 (12%)
Query 39 LEKNQCVFVAAHTSAGKTVVAEYAIALALQRRR------RCIFTSPLKALSNQKYRDFKQ 92
L N+ A T +GKTV+ E AI L R ++ +P+KAL +Q+Y ++KQ
Sbjct 200 LYSNKNFVACAPTGSGKTVLFELAIVRLLIEASEPWHNVRAVYMAPIKALCSQQYDNWKQ 259
Query 93 KFVSVGL----ITGDICIN 107
KF +GL +TGD I+
Sbjct 260 KFGPLGLKCKELTGDTEID 278
> tgo:TGME49_094350 DEAD/DEAH box helicase, putative
Length=2434
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 28/85 (32%), Positives = 43/85 (50%), Gaps = 18/85 (21%)
Query 26 FPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRRC------------ 73
F D +Q R + +++ + VFV A TS+GKT + YA+ L L RC
Sbjct 1350 FTPDYWQVRLLDLIDRRESVFVTAPTSSGKTFICFYAMELVL----RCPLRGEKAIDNDA 1405
Query 74 --IFTSPLKALSNQKYRDFKQKFVS 96
++ SP KAL++Q Y + +F S
Sbjct 1406 VIVYVSPSKALADQVYAEVHGRFSS 1430
> hsa:164045 HFM1, FLJ36760, FLJ39011, MER3, MGC163397, RP11-539G11.1,
SEC63D1, Si-11, Si-11-6, helicase; HFM1, ATP-dependent
DNA helicase homolog (S. cerevisiae) (EC:3.6.4.12)
Length=1435
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 25/71 (35%), Positives = 41/71 (57%), Gaps = 10/71 (14%)
Query 47 VAAHTSAGKTVVAEYAIALALQR------RRRCIFTSPLKALSNQKYRDFKQKFVSVGL- 99
+ A T +GKTVV E AI L + ++ +P+KAL +Q++ D+K+KF +GL
Sbjct 301 ICAPTGSGKTVVFELAITRLLMEVPLPWLNIKIVYMAPIKALCSQRFDDWKEKFGPIGLN 360
Query 100 ---ITGDICIN 107
+TGD ++
Sbjct 361 CKELTGDTVMD 371
> cpv:cgd2_2770 U5 small nuclear ribonucleoprotein 200kDA helicase,
Pre-mRNA splicing helicase BRR2 2 (RNA helicase plus Sec63
domain)
Length=2184
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 26/83 (31%), Positives = 47/83 (56%), Gaps = 22/83 (26%)
Query 42 NQCVFVAAHTSAGKTVVAEYAIALAL-----------------QRRRRCIFTSPLKALSN 84
++ +F+ A T +GKT+VAE AI AL +++ + ++ +PLK+L+N
Sbjct 1267 DENIFLGAPTGSGKTMVAEIAIFRALFADLDSKISISKLKPESKKKSKIVYIAPLKSLAN 1326
Query 85 QKYRDFKQKF-----VSVGLITG 102
+++ D+K F ++V LITG
Sbjct 1327 ERFNDWKFLFSNTLGLNVVLITG 1349
> sce:YGR271W SLH1; Putative RNA helicase related to Ski2p, involved
in translation inhibition of non-poly(A) mRNAs; required
for repressing propagation of dsRNA viruses (EC:3.6.1.-);
K01529 [EC:3.6.1.-]
Length=1967
Score = 42.0 bits (97), Expect = 6e-04, Method: Composition-based stats.
Identities = 30/96 (31%), Positives = 49/96 (51%), Gaps = 10/96 (10%)
Query 14 SPEEEPLAIEFGFPLDDFQKRAILRL----EKNQCVFVAAHTSAGKTVVAEYAIALALQR 69
S + PL IE +P F + N+ FV + T +GKT+VAE AI A +
Sbjct 1124 SALQNPL-IESIYPFKYFNPMQTMTFYTLYNTNENAFVGSPTGSGKTIVAELAIWHAFKT 1182
Query 70 --RRRCIFTSPLKALSNQKYRDFKQKFVSVGLITGD 103
++ ++ +P+KAL ++ D+++K V TGD
Sbjct 1183 FPGKKIVYIAPMKALVRERVDDWRKKITPV---TGD 1215
> tgo:TGME49_049810 activating signal cointegrator 1 complex subunit
3, putative (EC:5.99.1.3)
Length=2539
Score = 42.0 bits (97), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 35/55 (63%), Gaps = 2/55 (3%)
Query 42 NQCVFVAAHTSAGKTVVAEYAIA--LALQRRRRCIFTSPLKALSNQKYRDFKQKF 94
N V + A T +GKT+VAE A+ A +++ ++ +PLKAL+ ++ D+K +F
Sbjct 1564 NYNVLLGAPTGSGKTIVAELAMLRLFATSPKQKIVYIAPLKALAAERLEDWKARF 1618
> bbo:BBOV_I001530 19.m02171; helicase
Length=1798
Score = 41.6 bits (96), Expect = 7e-04, Method: Composition-based stats.
Identities = 26/65 (40%), Positives = 40/65 (61%), Gaps = 7/65 (10%)
Query 45 VFVAAHTSAGKTVVAEYAIALALQRRRRC---IFTSPLKALSNQKYRDFKQK---FVSVG 98
+ V A T +GKTVVAE A+ L R + C ++ +PLKAL+ ++ +D+ +K F V
Sbjct 1063 LLVGAPTGSGKTVVAELAM-FRLWRTQVCKKVVYIAPLKALAYERLKDWNKKFGMFKKVV 1121
Query 99 LITGD 103
+TGD
Sbjct 1122 EVTGD 1126
Score = 32.0 bits (71), Expect = 0.47, Method: Composition-based stats.
Identities = 22/79 (27%), Positives = 40/79 (50%), Gaps = 17/79 (21%)
Query 39 LEKNQCVFVAAHTSAGKTVVAEYAIALALQ----------RRRRCIFTSPLKALSNQKYR 88
+ +Q + ++A T GKT VA + ALQ + + ++ +P+KAL+++
Sbjct 181 FKTSQNMLISAPTGCGKTNVA---LLCALQNFESYFNGGEKNTKVVYVAPMKALASEVTG 237
Query 89 DFKQKFVSVGL----ITGD 103
F + V +GL +TGD
Sbjct 238 KFSKSLVDLGLRVREVTGD 256
> pfa:PF14_0370 DEAD/DEAH box helicase, putative; K01509 adenosinetriphosphatase
[EC:3.6.1.3]
Length=2472
Score = 41.6 bits (96), Expect = 7e-04, Method: Composition-based stats.
Identities = 19/58 (32%), Positives = 36/58 (62%), Gaps = 2/58 (3%)
Query 42 NQCVFVAAHTSAGKTVVAEYAIALALQR--RRRCIFTSPLKALSNQKYRDFKQKFVSV 97
++ + + A T +GKTV+ E I L R ++ ++ P+KA+ N++Y+ +K KF S+
Sbjct 1386 DENILLGAPTGSGKTVIGELCILRNLLRCEGQKSVYICPMKAIVNERYKSWKSKFKSL 1443
> ath:AT2G42270 U5 small nuclear ribonucleoprotein helicase, putative
Length=2172
Score = 41.2 bits (95), Expect = 8e-04, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 35/54 (64%), Gaps = 4/54 (7%)
Query 45 VFVAAHTSAGKTVVAEYAIAL----ALQRRRRCIFTSPLKALSNQKYRDFKQKF 94
V VAA T +GKT+ AE+AI R ++ +PL+A++ +++RD+++KF
Sbjct 1370 VVVAAPTGSGKTICAEFAILRNHLEGPDSAMRVVYIAPLEAIAKEQFRDWEKKF 1423
> cel:Y54E2A.6 hypothetical protein; K01529 [EC:3.6.1.-]
Length=1798
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 25/88 (28%), Positives = 49/88 (55%), Gaps = 10/88 (11%)
Query 40 EKNQCVFVAAHTSAGKTVVAEYAIALALQRR--RRCIFTSPLKALSNQKYRDFKQKF--- 94
+ ++ + A T +GKT+ AE A+ LQ + ++ +PLK+L ++ D+K+KF
Sbjct 974 KTDKSALIGAPTGSGKTLCAELAMFRLLQDHPGMKVVYIAPLKSLVRERVDDWKKKFEDG 1033
Query 95 --VSVGLITGDICINP---GAACLIVTT 117
V ++GD+ +P A+ +++TT
Sbjct 1034 MGYRVVEVSGDVTPDPEELAASSILITT 1061
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 19/74 (25%), Positives = 34/74 (45%), Gaps = 15/74 (20%)
Query 45 VFVAAHTSAGKTVVAEYAIALALQRRR-----------RCIFTSPLKALSNQKYRDFKQK 93
+ + A T AGKT +A I + + + I+ +P+KAL+ + F ++
Sbjct 111 LLICAPTGAGKTNIAMLTILNTIHEHQNSRGDIMKDDFKIIYIAPMKALATEMTESFGKR 170
Query 94 FVSVGL----ITGD 103
+GL +TGD
Sbjct 171 LAPLGLKVKELTGD 184
> tpv:TP01_0582 RNA helicase
Length=1764
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 22/70 (31%), Positives = 44/70 (62%), Gaps = 4/70 (5%)
Query 42 NQCVFVAAHTSAGKTVVAEYAIALALQR--RRRCIFTSPLKALSNQKYRDFKQK--FVSV 97
++ + VAA T +GKT+VAE + + + ++ +PLKAL++++++D+ +K F ++
Sbjct 1003 DESLVVAAPTGSGKTLVAELGLFRLFDKFPGKIAVYIAPLKALAHERFKDWCKKLHFKNI 1062
Query 98 GLITGDICIN 107
+TGD N
Sbjct 1063 LQLTGDTSSN 1072
> cel:C28H8.3 hypothetical protein; K01529 [EC:3.6.1.-]
Length=1714
Score = 39.3 bits (90), Expect = 0.004, Method: Composition-based stats.
Identities = 25/81 (30%), Positives = 41/81 (50%), Gaps = 6/81 (7%)
Query 25 GFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAIALALQRRRR--CIFTSPLKAL 82
GF D +Q++ + +++ + A TSAGKT V+ Y I L+ ++ +P KAL
Sbjct 782 GFAPDGWQRKMLDSVDRGNSALIIAPTSAGKTFVSYYCIEKVLRSSDNDVVVYVAPSKAL 841
Query 83 SNQ----KYRDFKQKFVSVGL 99
NQ Y F+ K + G+
Sbjct 842 INQVCGSVYARFRNKSMKRGI 862
> ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding /
ATP-dependent helicase/ helicase/ nucleic acid binding / nucleoside-triphosphatase/
nucleotide binding; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2171
Score = 38.1 bits (87), Expect = 0.007, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 34/57 (59%), Gaps = 4/57 (7%)
Query 42 NQCVFVAAHTSAGKTVVAEYAIALALQR----RRRCIFTSPLKALSNQKYRDFKQKF 94
N V VAA T +GKT+ AE+AI R ++ +PL+A++ +++R ++ KF
Sbjct 1366 NDNVLVAAPTGSGKTICAEFAILRNHHEGPDATMRVVYIAPLEAIAKEQFRIWEGKF 1422
> bbo:BBOV_III000950 17.m07111; sec63 domain containing protein
(EC:3.6.1.-); K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2133
Score = 37.7 bits (86), Expect = 0.011, Method: Composition-based stats.
Identities = 25/77 (32%), Positives = 40/77 (51%), Gaps = 15/77 (19%)
Query 45 VFVAAHTSAGKTVVAEYAIALALQRRR-----------RCIFTSPLKALSNQKYRDFKQK 93
+ V A T AGKT VA A+ L R R + ++ SP+K+L ++ + F Q+
Sbjct 501 MLVCAPTGAGKTNVAVLAMLSVLDRHRDETGNLNLHDFKIVYISPMKSLVMEQAQSFSQR 560
Query 94 F----VSVGLITGDICI 106
F +SV +TGD+ +
Sbjct 561 FAPYGISVRELTGDMSL 577
> tgo:TGME49_023390 sec63 domain-containing DEAD/DEAH box helicase,
putative ; K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2198
Score = 37.4 bits (85), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 42/83 (50%), Gaps = 16/83 (19%)
Query 40 EKNQCVFVAAHTSAGKTVVAEYAIALALQRRR------------RCIFTSPLKALSNQKY 87
E ++ + + A T AGKT VA AI + R R + ++ SP+KAL ++
Sbjct 547 EFHENLLLCAPTGAGKTNVAMLAILNVIGRHRNAKTGAVDLASFKVVYISPMKALVAEQV 606
Query 88 RDFKQKF----VSVGLITGDICI 106
+ F Q+ VSV +TGD+ +
Sbjct 607 QAFSQRLQPYGVSVRELTGDVNL 629
> sce:YGL251C HFM1, MER3; Hfm1p (EC:3.6.1.-); K01529 [EC:3.6.1.-]
Length=1187
Score = 36.6 bits (83), Expect = 0.024, Method: Composition-based stats.
Identities = 22/72 (30%), Positives = 40/72 (55%), Gaps = 8/72 (11%)
Query 40 EKNQCVFVAAHTSAGKTVVAEYAIALALQR------RRRCIFTSPLKALSNQKYRDFKQK 93
E N+ +++ T +GKTV+ E AI ++ + I+ +P K+L + Y+++
Sbjct 152 ESNENCIISSPTGSGKTVLFELAILRLIKETNSDTNNTKIIYIAPTKSLCYEMYKNWFPS 211
Query 94 FV--SVGLITGD 103
FV SVG++T D
Sbjct 212 FVNLSVGMLTSD 223
> ath:AT5G61140 DEAD box RNA helicase, putative; K01529 [EC:3.6.1.-]
Length=2146
Score = 36.2 bits (82), Expect = 0.027, Method: Composition-based stats.
Identities = 18/54 (33%), Positives = 32/54 (59%), Gaps = 2/54 (3%)
Query 45 VFVAAHTSAGKTVVAEYAIA--LALQRRRRCIFTSPLKALSNQKYRDFKQKFVS 96
V V A T +GKT+ AE A+ + Q + ++ +PLKA+ ++ D+K+ V+
Sbjct 1363 VLVGAPTGSGKTISAELAMLRLFSTQPDMKVVYIAPLKAIVRERMNDWKKHLVA 1416
> cpv:cgd5_2070 SPAC694.02. SKI family SFII helicase
Length=2123
Score = 35.8 bits (81), Expect = 0.034, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 37/73 (50%), Gaps = 2/73 (2%)
Query 26 FPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAI--ALALQRRRRCIFTSPLKALS 83
F D +QK+ + ++ V A T++GKT + YA+ L +F +P KAL+
Sbjct 1052 FSPDYWQKQILDIIDSGDSALVCAPTASGKTFICYYAMEQVLRFDNESVVVFIAPTKALA 1111
Query 84 NQKYRDFKQKFVS 96
+Q + + +F S
Sbjct 1112 DQVHAEIAYRFGS 1124
> hsa:55601 DDX60, FLJ10787, FLJ20035; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 60 (EC:3.6.4.13)
Length=1712
Score = 35.8 bits (81), Expect = 0.039, Method: Composition-based stats.
Identities = 26/106 (24%), Positives = 47/106 (44%), Gaps = 16/106 (15%)
Query 1 DEEEEEAAATQDLSPEEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAE 60
DE ++ QD P D +Q+ + ++KN+ + A TS+GKT +
Sbjct 750 DERKDPDPRVQDFIP-------------DTWQRELLDVVDKNESAVIVAPTSSGKTYASY 796
Query 61 YAIALALQRRRR--CIFTSPLKALSNQKYRDFKQKFVSVGLITGDI 104
Y + L+ ++ +P KAL NQ + +F L +G++
Sbjct 797 YCMEKVLKESDDGVVVYVAPTKALVNQVAATVQNRFTK-NLPSGEV 841
> mmu:71602 Myo1e, 2310020N23Rik, 9130023P14Rik, AA407778, myosin-1e,
myr_3; myosin IE; K10356 myosin I
Length=1107
Score = 35.4 bits (80), Expect = 0.045, Method: Composition-based stats.
Identities = 20/61 (32%), Positives = 32/61 (52%), Gaps = 3/61 (4%)
Query 4 EEEAAATQDLSPEEEPLAIEFGFPLDDFQKRAILRLEKNQCVFVAAHTSAGKTVVAEYAI 63
E+E Q + E P I + L D R ++ +NQCV ++ + AGKTV A+Y +
Sbjct 70 EKEVEMYQGAAQYENPPHI---YALADSMYRNMIIDRENQCVIISGESGAGKTVAAKYIM 126
Query 64 A 64
+
Sbjct 127 S 127
Lambda K H
0.322 0.137 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2231140792
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40