bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
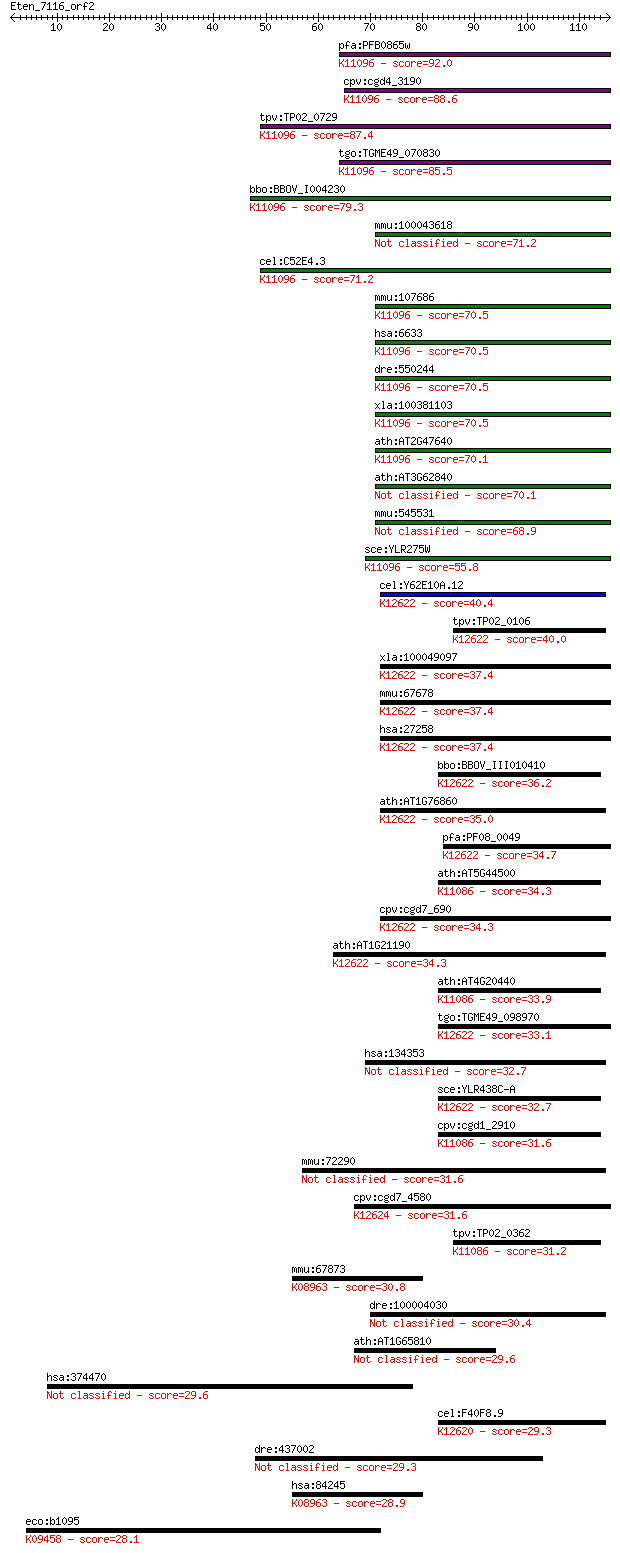
Query= Eten_7116_orf2
Length=115
Score E
Sequences producing significant alignments: (Bits) Value
pfa:PFB0865w small nuclear ribonucleoprotein, putative; K11096... 92.0 4e-19
cpv:cgd4_3190 small nuclear ribonucleoprotein ; K11096 small n... 88.6 5e-18
tpv:TP02_0729 small nuclear ribonucleoprotein D2; K11096 small... 87.4 9e-18
tgo:TGME49_070830 small nuclear ribonucleoprotein Sm D2, putat... 85.5 3e-17
bbo:BBOV_I004230 19.m02306; small nuclear ribonucleoprotein; K... 79.3 2e-15
mmu:100043618 Gm10120; small nuclear ribonucleoprotein D2 pseu... 71.2 8e-13
cel:C52E4.3 snr-4; Small Nuclear Ribonucleoprotein family memb... 71.2 8e-13
mmu:107686 Snrpd2, 1810009A06Rik, MGC62953, SMD2; small nuclea... 70.5 1e-12
hsa:6633 SNRPD2, SMD2, SNRPD1, Sm-D2; small nuclear ribonucleo... 70.5 1e-12
dre:550244 snrpd2, im:6908977, si:dkey-113g17.2, zgc:110732; s... 70.5 1e-12
xla:100381103 snrpd2, sm-d2, smd2, snrpd1; small nuclear ribon... 70.5 1e-12
ath:AT2G47640 small nuclear ribonucleoprotein D2, putative / s... 70.1 2e-12
ath:AT3G62840 hypothetical protein 70.1 2e-12
mmu:545531 Gm5848, EG545531; predicted pseudogene 5848 68.9
sce:YLR275W SMD2; Sm D2; K11096 small nuclear ribonucleoprotei... 55.8 4e-08
cel:Y62E10A.12 lsm-3; LSM Sm-like protein family member (lsm-3... 40.4 0.002
tpv:TP02_0106 hypothetical protein; K12622 U6 snRNA-associated... 40.0 0.002
xla:100049097 lsm3; LSM3 homolog, U6 small nuclear RNA associa... 37.4 0.011
mmu:67678 Lsm3, 1010001J12Rik, 2610005D18Rik, 6030401D18Rik, S... 37.4 0.013
hsa:27258 LSM3, SMX4, USS2, YLR438C; LSM3 homolog, U6 small nu... 37.4 0.013
bbo:BBOV_III010410 17.m07899; LSM domain containing protein; K... 36.2 0.029
ath:AT1G76860 small nuclear ribonucleoprotein, putative / snRN... 35.0 0.059
pfa:PF08_0049 lsm3 homologue, putative; K12622 U6 snRNA-associ... 34.7 0.080
ath:AT5G44500 small nuclear ribonucleoprotein associated prote... 34.3 0.090
cpv:cgd7_690 small nuclear ribonucleoprotein ; K12622 U6 snRNA... 34.3 0.091
ath:AT1G21190 small nuclear ribonucleoprotein, putative / snRN... 34.3 0.095
ath:AT4G20440 smB; smB (small nuclear ribonucleoprotein associ... 33.9 0.12
tgo:TGME49_098970 LSM domain-containing protein ; K12622 U6 sn... 33.1 0.23
hsa:134353 LSM11, FLJ38273; LSM11, U7 small nuclear RNA associ... 32.7 0.26
sce:YLR438C-A LSM3, SMX4, USS2; Lsm (Like Sm) protein; part of... 32.7 0.30
cpv:cgd1_2910 hypothetical protein ; K11086 small nuclear ribo... 31.6 0.62
mmu:72290 Lsm11, 2210404M20Rik, AV079530; U7 snRNP-specific Sm... 31.6 0.64
cpv:cgd7_4580 U6 snRNA-associated Sm-like protein LSm5. SM dom... 31.6 0.66
tpv:TP02_0362 small nuclear ribonucleoprotein B; K11086 small ... 31.2 0.89
mmu:67873 Mri1, 2410018C20Rik; methylthioribose-1-phosphate is... 30.8 1.0
dre:100004030 lsm11, im:7149708; LSM11, U7 small nuclear RNA a... 30.4 1.5
ath:AT1G65810 tRNA-splicing endonuclease positive effector-rel... 29.6 2.3
hsa:374470 C12orf42, FLJ25323, MGC43592, MGC57409; chromosome ... 29.6 2.5
cel:F40F8.9 lsm-1; LSM Sm-like protein family member (lsm-1); ... 29.3 2.8
dre:437002 wipi2, zgc:92576; WD repeat domain, phosphoinositid... 29.3 3.6
hsa:84245 MRI1, MGC3207, MTNA, Ypr118w; methylthioribose-1-pho... 28.9 4.3
eco:b1095 fabF, cvc, ECK1081, fabJ, JW1081, vtr; 3-oxoacyl-[ac... 28.1 7.4
> pfa:PFB0865w small nuclear ribonucleoprotein, putative; K11096
small nuclear ribonucleoprotein D2
Length=101
Score = 92.0 bits (227), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 43/52 (82%), Positives = 46/52 (88%), Gaps = 0/52 (0%)
Query 64 NRDNPEGGPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVR 115
NRDNPE GPL LLS CV DN+QVL+NCRNNRK+L RVKAFDRHCNLLLT VR
Sbjct 10 NRDNPEDGPLGLLSECVKDNAQVLINCRNNRKILGRVKAFDRHCNLLLTGVR 61
> cpv:cgd4_3190 small nuclear ribonucleoprotein ; K11096 small
nuclear ribonucleoprotein D2
Length=106
Score = 88.6 bits (218), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 38/51 (74%), Positives = 44/51 (86%), Gaps = 0/51 (0%)
Query 65 RDNPEGGPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVR 115
+DNP GPL+LL+ CV N+Q+LVNCRNNRK+L RVKAFDRHCNLLLTD R
Sbjct 14 KDNPSEGPLSLLTECVKTNTQILVNCRNNRKILGRVKAFDRHCNLLLTDAR 64
> tpv:TP02_0729 small nuclear ribonucleoprotein D2; K11096 small
nuclear ribonucleoprotein D2
Length=106
Score = 87.4 bits (215), Expect = 9e-18, Method: Compositional matrix adjust.
Identities = 40/67 (59%), Positives = 50/67 (74%), Gaps = 1/67 (1%)
Query 49 MAAAADGDKAAPHLVNRDNPEGGPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCN 108
M+ D L N+DNP GPL++L CV +N QVL+NCRNNRK+LARVKAFDRHCN
Sbjct 1 MSTKVKQDPTPMELDNKDNP-AGPLSILEECVRENCQVLINCRNNRKILARVKAFDRHCN 59
Query 109 LLLTDVR 115
++LT+VR
Sbjct 60 MILTNVR 66
> tgo:TGME49_070830 small nuclear ribonucleoprotein Sm D2, putative
; K11096 small nuclear ribonucleoprotein D2
Length=104
Score = 85.5 bits (210), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 39/52 (75%), Positives = 46/52 (88%), Gaps = 0/52 (0%)
Query 64 NRDNPEGGPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVR 115
NRD+P GPL++L+ V DNSQVL+NCRNNRK+L RVKAFDRHCNL+LTDVR
Sbjct 11 NRDHPPEGPLSVLAASVKDNSQVLINCRNNRKVLGRVKAFDRHCNLVLTDVR 62
> bbo:BBOV_I004230 19.m02306; small nuclear ribonucleoprotein;
K11096 small nuclear ribonucleoprotein D2
Length=162
Score = 79.3 bits (194), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 39/69 (56%), Positives = 47/69 (68%), Gaps = 9/69 (13%)
Query 47 GIMAAAADGDKAAPHLVNRDNPEGGPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRH 106
GI + + DK P GPL++L CV DNSQVL+NCR+NRKLL RVKAFDRH
Sbjct 63 GIRSMSTTEDKDCP---------SGPLSVLEACVRDNSQVLINCRSNRKLLGRVKAFDRH 113
Query 107 CNLLLTDVR 115
N++LTDVR
Sbjct 114 FNMILTDVR 122
> mmu:100043618 Gm10120; small nuclear ribonucleoprotein D2 pseudogene
Length=118
Score = 71.2 bits (173), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 29/45 (64%), Positives = 41/45 (91%), Gaps = 0/45 (0%)
Query 71 GPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVR 115
GPL++L++ V +N+QVL+NCRNN+KLL RVKAFDRHCN++L +V+
Sbjct 27 GPLSVLTQSVKNNTQVLINCRNNKKLLGRVKAFDRHCNMVLENVK 71
> cel:C52E4.3 snr-4; Small Nuclear Ribonucleoprotein family member
(snr-4); K11096 small nuclear ribonucleoprotein D2
Length=118
Score = 71.2 bits (173), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 35/71 (49%), Positives = 50/71 (70%), Gaps = 4/71 (5%)
Query 49 MAAAAD--GDKAAPHLVNRDNPEG--GPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFD 104
M+A A + A L +++ E GPL++L+ V +N QVL+NCRNN+KLL RVKAFD
Sbjct 1 MSAQAKPRSEMTAEELAAKEDEEFNVGPLSILTNSVKNNHQVLINCRNNKKLLGRVKAFD 60
Query 105 RHCNLLLTDVR 115
RHCN++L +V+
Sbjct 61 RHCNMVLENVK 71
> mmu:107686 Snrpd2, 1810009A06Rik, MGC62953, SMD2; small nuclear
ribonucleoprotein D2; K11096 small nuclear ribonucleoprotein
D2
Length=118
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 29/45 (64%), Positives = 41/45 (91%), Gaps = 0/45 (0%)
Query 71 GPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVR 115
GPL++L++ V +N+QVL+NCRNN+KLL RVKAFDRHCN++L +V+
Sbjct 27 GPLSVLTQSVKNNTQVLINCRNNKKLLGRVKAFDRHCNMVLENVK 71
> hsa:6633 SNRPD2, SMD2, SNRPD1, Sm-D2; small nuclear ribonucleoprotein
D2 polypeptide 16.5kDa; K11096 small nuclear ribonucleoprotein
D2
Length=118
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 29/45 (64%), Positives = 41/45 (91%), Gaps = 0/45 (0%)
Query 71 GPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVR 115
GPL++L++ V +N+QVL+NCRNN+KLL RVKAFDRHCN++L +V+
Sbjct 27 GPLSVLTQSVKNNTQVLINCRNNKKLLGRVKAFDRHCNMVLENVK 71
> dre:550244 snrpd2, im:6908977, si:dkey-113g17.2, zgc:110732;
small nuclear ribonucleoprotein D2 polypeptide; K11096 small
nuclear ribonucleoprotein D2
Length=118
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 29/45 (64%), Positives = 41/45 (91%), Gaps = 0/45 (0%)
Query 71 GPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVR 115
GPL++L++ V +N+QVL+NCRNN+KLL RVKAFDRHCN++L +V+
Sbjct 27 GPLSVLTQSVKNNTQVLINCRNNKKLLGRVKAFDRHCNMVLENVK 71
> xla:100381103 snrpd2, sm-d2, smd2, snrpd1; small nuclear ribonucleoprotein
D2 polypeptide 16.5kDa; K11096 small nuclear
ribonucleoprotein D2
Length=118
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 29/45 (64%), Positives = 41/45 (91%), Gaps = 0/45 (0%)
Query 71 GPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVR 115
GPL++L++ V +N+QVL+NCRNN+KLL RVKAFDRHCN++L +V+
Sbjct 27 GPLSVLTQSVKNNTQVLINCRNNKKLLGRVKAFDRHCNMVLENVK 71
> ath:AT2G47640 small nuclear ribonucleoprotein D2, putative /
snRNP core protein D2, putative / Sm protein D2, putative;
K11096 small nuclear ribonucleoprotein D2
Length=109
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 30/45 (66%), Positives = 39/45 (86%), Gaps = 0/45 (0%)
Query 71 GPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVR 115
GPL++L V +N+QVL+NCRNNRKLL RV+AFDRHCN++L +VR
Sbjct 22 GPLSVLMMSVKNNTQVLINCRNNRKLLGRVRAFDRHCNMVLENVR 66
> ath:AT3G62840 hypothetical protein
Length=108
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 30/45 (66%), Positives = 39/45 (86%), Gaps = 0/45 (0%)
Query 71 GPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVR 115
GPL++L V +N+QVL+NCRNNRKLL RV+AFDRHCN++L +VR
Sbjct 21 GPLSVLMMSVKNNTQVLINCRNNRKLLGRVRAFDRHCNMVLENVR 65
> mmu:545531 Gm5848, EG545531; predicted pseudogene 5848
Length=118
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 29/45 (64%), Positives = 40/45 (88%), Gaps = 0/45 (0%)
Query 71 GPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVR 115
GPL++LS+ V +N+QVL+NCRN +KLL RVKAFDRHCN++L +V+
Sbjct 27 GPLSVLSQSVKNNTQVLINCRNKKKLLGRVKAFDRHCNMVLENVK 71
> sce:YLR275W SMD2; Sm D2; K11096 small nuclear ribonucleoprotein
D2
Length=110
Score = 55.8 bits (133), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 21/47 (44%), Positives = 38/47 (80%), Gaps = 0/47 (0%)
Query 69 EGGPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVR 115
+ GP++L++ ++ + V+++ RNN K++ARVKAFDRHCN++L +V+
Sbjct 27 KHGPMSLINDAMVTRTPVIISLRNNHKIIARVKAFDRHCNMVLENVK 73
> cel:Y62E10A.12 lsm-3; LSM Sm-like protein family member (lsm-3);
K12622 U6 snRNA-associated Sm-like protein LSm3
Length=102
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 21/43 (48%), Positives = 31/43 (72%), Gaps = 2/43 (4%)
Query 72 PLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDV 114
PL LL R LD +V V RN+R+L R++AFD+H N++L++V
Sbjct 17 PLDLL-RLSLDE-RVYVKMRNDRELRGRLRAFDQHLNMVLSEV 57
> tpv:TP02_0106 hypothetical protein; K12622 U6 snRNA-associated
Sm-like protein LSm3
Length=84
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 24/29 (82%), Gaps = 0/29 (0%)
Query 86 VLVNCRNNRKLLARVKAFDRHCNLLLTDV 114
+ + C+N R+L+ R++AFD HCN++L++V
Sbjct 22 IYLKCKNGRELVGRLQAFDEHCNMVLSEV 50
> xla:100049097 lsm3; LSM3 homolog, U6 small nuclear RNA associated;
K12622 U6 snRNA-associated Sm-like protein LSm3
Length=102
Score = 37.4 bits (85), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 19/44 (43%), Positives = 29/44 (65%), Gaps = 2/44 (4%)
Query 72 PLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVR 115
PL L+ R LD ++ V RN+R+L R+ A+D+H N++L DV
Sbjct 17 PLDLI-RLSLD-ERIYVKMRNDRELRGRLNAYDQHLNMILGDVE 58
> mmu:67678 Lsm3, 1010001J12Rik, 2610005D18Rik, 6030401D18Rik,
SMX4, USS2; LSM3 homolog, U6 small nuclear RNA associated (S.
cerevisiae); K12622 U6 snRNA-associated Sm-like protein LSm3
Length=102
Score = 37.4 bits (85), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 19/44 (43%), Positives = 29/44 (65%), Gaps = 2/44 (4%)
Query 72 PLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVR 115
PL L+ R LD ++ V RN+R+L R+ A+D+H N++L DV
Sbjct 17 PLDLI-RLSLD-ERIYVKMRNDRELRGRLHAYDQHLNMILGDVE 58
> hsa:27258 LSM3, SMX4, USS2, YLR438C; LSM3 homolog, U6 small
nuclear RNA associated (S. cerevisiae); K12622 U6 snRNA-associated
Sm-like protein LSm3
Length=102
Score = 37.4 bits (85), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 19/44 (43%), Positives = 29/44 (65%), Gaps = 2/44 (4%)
Query 72 PLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVR 115
PL L+ R LD ++ V RN+R+L R+ A+D+H N++L DV
Sbjct 17 PLDLI-RLSLD-ERIYVKMRNDRELRGRLHAYDQHLNMILGDVE 58
> bbo:BBOV_III010410 17.m07899; LSM domain containing protein;
K12622 U6 snRNA-associated Sm-like protein LSm3
Length=89
Score = 36.2 bits (82), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 11/31 (35%), Positives = 23/31 (74%), Gaps = 0/31 (0%)
Query 83 NSQVLVNCRNNRKLLARVKAFDRHCNLLLTD 113
+ +V + C+ R+++ ++ A+D HCN+LL+D
Sbjct 14 DERVYLKCKGGREIVGQLHAYDEHCNMLLSD 44
> ath:AT1G76860 small nuclear ribonucleoprotein, putative / snRNP,
putative / Sm protein, putative; K12622 U6 snRNA-associated
Sm-like protein LSm3
Length=98
Score = 35.0 bits (79), Expect = 0.059, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 29/43 (67%), Gaps = 2/43 (4%)
Query 72 PLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDV 114
PL L+ R LD ++ V R++R+L ++ AFD+H N++L DV
Sbjct 12 PLDLI-RLSLDE-RIYVKLRSDRELRGKLHAFDQHLNMILGDV 52
> pfa:PF08_0049 lsm3 homologue, putative; K12622 U6 snRNA-associated
Sm-like protein LSm3
Length=91
Score = 34.7 bits (78), Expect = 0.080, Method: Compositional matrix adjust.
Identities = 10/32 (31%), Positives = 24/32 (75%), Gaps = 0/32 (0%)
Query 84 SQVLVNCRNNRKLLARVKAFDRHCNLLLTDVR 115
++ + C+ +R+L+ ++ A+D H N++L++VR
Sbjct 20 EEIFLKCKGDRELIGKLDAYDNHLNMILSNVR 51
> ath:AT5G44500 small nuclear ribonucleoprotein associated protein
B, putative / snRNP-B, putative / Sm protein B, putative;
K11086 small nuclear ribonucleoprotein B and B'
Length=254
Score = 34.3 bits (77), Expect = 0.090, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 83 NSQVLVNCRNNRKLLARVKAFDRHCNLLLTD 113
N ++ V ++ R+L+ + AFDRH NL+L D
Sbjct 14 NYRMRVTIQDGRQLIGKFMAFDRHMNLVLGD 44
> cpv:cgd7_690 small nuclear ribonucleoprotein ; K12622 U6 snRNA-associated
Sm-like protein LSm3
Length=95
Score = 34.3 bits (77), Expect = 0.091, Method: Compositional matrix adjust.
Identities = 20/44 (45%), Positives = 27/44 (61%), Gaps = 2/44 (4%)
Query 72 PLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVR 115
PL L+ R LD QV V CR NR+L + A+D H N++L +V
Sbjct 7 PLDLI-RLSLD-EQVFVKCRGNRELKGTLYAYDPHMNMVLGNVE 48
> ath:AT1G21190 small nuclear ribonucleoprotein, putative / snRNP,
putative / Sm protein, putative; K12622 U6 snRNA-associated
Sm-like protein LSm3
Length=97
Score = 34.3 bits (77), Expect = 0.095, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 29/52 (55%), Gaps = 2/52 (3%)
Query 63 VNRDNPEGGPLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDV 114
V D PL L+ + ++ V R++R+L ++ AFD+H N++L DV
Sbjct 3 VEEDATVREPLDLIRLSI--EERIYVKLRSDRELRGKLHAFDQHLNMILGDV 52
> ath:AT4G20440 smB; smB (small nuclear ribonucleoprotein associated
protein B); K11086 small nuclear ribonucleoprotein B
and B'
Length=257
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 83 NSQVLVNCRNNRKLLARVKAFDRHCNLLLTD 113
N ++ V ++ R+L+ + AFDRH NL+L D
Sbjct 14 NYRMRVTIQDGRQLVGKFMAFDRHMNLVLGD 44
> tgo:TGME49_098970 LSM domain-containing protein ; K12622 U6
snRNA-associated Sm-like protein LSm3
Length=96
Score = 33.1 bits (74), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 11/33 (33%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 83 NSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVR 115
+ +V + CR +R+++ ++ A+D H N++L DV
Sbjct 19 DERVTIKCRGDREVVGKLHAYDMHLNMVLGDVE 51
> hsa:134353 LSM11, FLJ38273; LSM11, U7 small nuclear RNA associated
Length=360
Score = 32.7 bits (73), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 27/50 (54%), Gaps = 4/50 (8%)
Query 69 EGGPLTLLSRCVLDNSQVLVNCRNNRKL----LARVKAFDRHCNLLLTDV 114
EG PL L RC+ + +V V+ R + L + AFD+ N+ LTDV
Sbjct 152 EGSPLGELHRCIREGVKVNVHIRTFKGLRGVCTGFLVAFDKFWNMALTDV 201
> sce:YLR438C-A LSM3, SMX4, USS2; Lsm (Like Sm) protein; part
of heteroheptameric complexes (Lsm2p-7p and either Lsm1p or
8p): cytoplasmic Lsm1p complex involved in mRNA decay; nuclear
Lsm8p complex part of U6 snRNP and possibly involved in processing
tRNA, snoRNA, and rRNA; K12622 U6 snRNA-associated
Sm-like protein LSm3
Length=89
Score = 32.7 bits (73), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 12/31 (38%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 83 NSQVLVNCRNNRKLLARVKAFDRHCNLLLTD 113
+ +V + R R L+ ++AFD HCN++L+D
Sbjct 13 DERVYIKLRGARTLVGTLQAFDSHCNIVLSD 43
> cpv:cgd1_2910 hypothetical protein ; K11086 small nuclear ribonucleoprotein
B and B'
Length=139
Score = 31.6 bits (70), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 12/31 (38%), Positives = 22/31 (70%), Gaps = 0/31 (0%)
Query 83 NSQVLVNCRNNRKLLARVKAFDRHCNLLLTD 113
N ++ V +++R ++ + AFDRH NL+L+D
Sbjct 6 NYRIRVTVQDDRVMVGNLMAFDRHMNLVLSD 36
> mmu:72290 Lsm11, 2210404M20Rik, AV079530; U7 snRNP-specific
Sm-like protein LSM11
Length=361
Score = 31.6 bits (70), Expect = 0.64, Method: Composition-based stats.
Identities = 24/64 (37%), Positives = 32/64 (50%), Gaps = 6/64 (9%)
Query 57 KAAPHLVNRDNP--EGGPLTLLSRCVLDNSQVLVNCRNNRKL----LARVKAFDRHCNLL 110
K AP V P EG PL L RC+ + +V V+ R + L + AFD+ N+
Sbjct 139 KKAPRNVLTRMPLHEGSPLGELHRCIREGVKVNVHIRTFKGLRGVCTGFLVAFDKFWNMA 198
Query 111 LTDV 114
LTDV
Sbjct 199 LTDV 202
> cpv:cgd7_4580 U6 snRNA-associated Sm-like protein LSm5. SM domain
; K12624 U6 snRNA-associated Sm-like protein LSm5
Length=121
Score = 31.6 bits (70), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 14/53 (26%), Positives = 31/53 (58%), Gaps = 6/53 (11%)
Query 67 NPEGG----PLTLLSRCVLDNSQVLVNCRNNRKLLARVKAFDRHCNLLLTDVR 115
N +GG PL L+ +C+ +++ V + +++ ++ FD + N++L DV+
Sbjct 20 NQKGGNIILPLALIDKCI--GNRIYVVMKGDKEFSGVLRGFDEYVNMVLDDVQ 70
> tpv:TP02_0362 small nuclear ribonucleoprotein B; K11086 small
nuclear ribonucleoprotein B and B'
Length=155
Score = 31.2 bits (69), Expect = 0.89, Method: Compositional matrix adjust.
Identities = 11/28 (39%), Positives = 20/28 (71%), Gaps = 0/28 (0%)
Query 86 VLVNCRNNRKLLARVKAFDRHCNLLLTD 113
V V ++NRK + + A+D++ NL+L+D
Sbjct 16 VRVTLKDNRKFVGTLVAYDKYMNLVLSD 43
> mmu:67873 Mri1, 2410018C20Rik; methylthioribose-1-phosphate
isomerase homolog (S. cerevisiae) (EC:5.3.1.23); K08963 methylthioribose-1-phosphate
isomerase [EC:5.3.1.23]
Length=369
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 55 GDKAAPHLVNRDNPEGGPLTLLSRC 79
GD A HL+ + NP GG +T+L+ C
Sbjct 144 GDLGARHLLEQTNPRGGKVTVLTHC 168
> dre:100004030 lsm11, im:7149708; LSM11, U7 small nuclear RNA
associated
Length=363
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 19/49 (38%), Positives = 26/49 (53%), Gaps = 4/49 (8%)
Query 70 GGPLTLLSRCVLDNSQVLVNCRNNRKLLAR----VKAFDRHCNLLLTDV 114
G PL L+RCV + +V V+ R + L V AFD+ NL + DV
Sbjct 189 GSPLGQLNRCVQEKIRVKVHIRTFKGLRGVCSGFVVAFDKFWNLAMVDV 237
> ath:AT1G65810 tRNA-splicing endonuclease positive effector-related
Length=1050
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 14/31 (45%), Positives = 20/31 (64%), Gaps = 4/31 (12%)
Query 67 NPEGGPLTLLSRCVLDNSQV----LVNCRNN 93
NPEGG L L+SR + N++V V+C+ N
Sbjct 219 NPEGGNLKLISRVLQSNNEVDGGSCVSCKEN 249
> hsa:374470 C12orf42, FLJ25323, MGC43592, MGC57409; chromosome
12 open reading frame 42
Length=360
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 29/85 (34%), Positives = 36/85 (42%), Gaps = 15/85 (17%)
Query 8 LASTGPAAAA----GQACMQLGRQA-PSIGLGGLLGASGTFVGKGIMAAAADGDKAAPHL 62
L STGP+ + + G QA P LLGASG VGKG +A A + PH
Sbjct 232 LQSTGPSNTELEPEERMAVPAGAQAHPDDIQSRLLGASGNPVGKGAVAMAPEMLPKHPHT 291
Query 63 VNRDNPE----------GGPLTLLS 77
P+ G PL LL+
Sbjct 292 PRDRRPQADTSLHGNLAGAPLPLLA 316
> cel:F40F8.9 lsm-1; LSM Sm-like protein family member (lsm-1);
K12620 U6 snRNA-associated Sm-like protein LSm1
Length=125
Score = 29.3 bits (64), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 12/32 (37%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 83 NSQVLVNCRNNRKLLARVKAFDRHCNLLLTDV 114
+ ++LV R+ RKL+ +++ D+ NL+L DV
Sbjct 19 DKKLLVVLRDGRKLIGFLRSIDQFANLILEDV 50
> dre:437002 wipi2, zgc:92576; WD repeat domain, phosphoinositide
interacting 2
Length=131
Score = 29.3 bits (64), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 32/60 (53%), Gaps = 8/60 (13%)
Query 48 IMAAAADGDKAAPHLVNRDNPEGGPLTLLSRCVLDNS-----QVLVNCRNNRKLLARVKA 102
++ AAADG +L N D EGG TL+ + LD S ++L ++R L+A+ +
Sbjct 33 LLVAAADG---YLYLYNLDPQEGGECTLMKQHKLDGSAEPANEILEQTAHDRPLVAQTYS 89
> hsa:84245 MRI1, MGC3207, MTNA, Ypr118w; methylthioribose-1-phosphate
isomerase homolog (S. cerevisiae) (EC:5.3.1.23); K08963
methylthioribose-1-phosphate isomerase [EC:5.3.1.23]
Length=369
Score = 28.9 bits (63), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 55 GDKAAPHLVNRDNPEGGPLTLLSRC 79
GD A HL+ R P GG +T+L+ C
Sbjct 144 GDLGARHLLERVAPSGGKVTVLTHC 168
> eco:b1095 fabF, cvc, ECK1081, fabJ, JW1081, vtr; 3-oxoacyl-[acyl-carrier-protein]
synthase II (EC:2.3.1.41); K09458 3-oxoacyl-[acyl-carrier-protein]
synthase II [EC:2.3.1.179]
Length=413
Score = 28.1 bits (61), Expect = 7.4, Method: Composition-based stats.
Identities = 25/78 (32%), Positives = 37/78 (47%), Gaps = 12/78 (15%)
Query 4 FLNPLASTGPAA--AAGQACMQLGRQAPSIGL--------GGLLGASGTFVGKGIMAAAA 53
++N ++ PA A QA + +A S L G LLGA+G + I + A
Sbjct 300 YVNAHGTSTPAGDKAEAQAVKTIFGEAASRVLVSSTKSMTGHLLGAAGAV--ESIYSILA 357
Query 54 DGDKAAPHLVNRDNPEGG 71
D+A P +N DNP+ G
Sbjct 358 LRDQAVPPTINLDNPDEG 375
Lambda K H
0.321 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2042676840
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40