bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7145_orf2
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
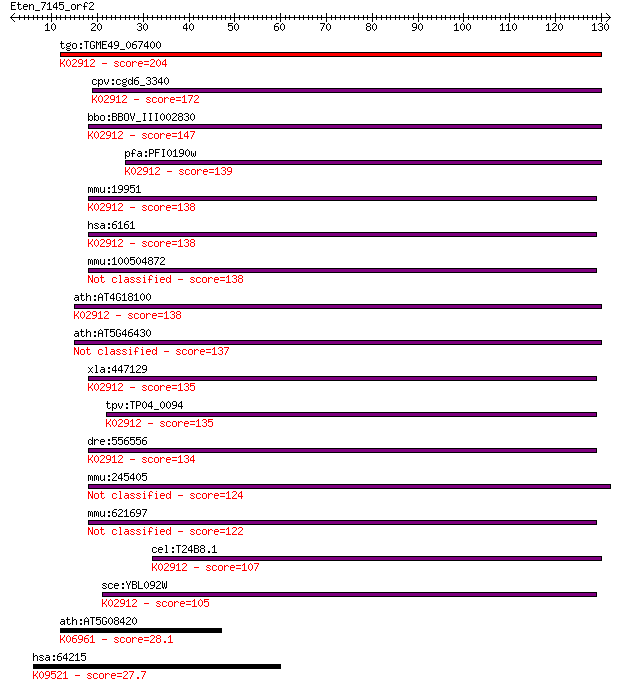
tgo:TGME49_067400 ribosomal protein L32, putative ; K02912 lar... 204 5e-53
cpv:cgd6_3340 60S ribosomal protein L32 ; K02912 large subunit... 172 3e-43
bbo:BBOV_III002830 17.m07268; 60S ribosomal protein L32; K0291... 147 6e-36
pfa:PFI0190w 60S ribosomal protein L32, putative; K02912 large... 139 2e-33
mmu:19951 Rpl32, AU020185, rpL32-3A; ribosomal protein L32; K0... 138 4e-33
hsa:6161 RPL32; ribosomal protein L32; K02912 large subunit ri... 138 4e-33
mmu:100504872 60S ribosomal protein L32-like 138 4e-33
ath:AT4G18100 60S ribosomal protein L32 (RPL32A); K02912 large... 138 4e-33
ath:AT5G46430 60S ribosomal protein L32 (RPL32B) 137 7e-33
xla:447129 rpl32, MGC85374, MGC86311; ribosomal protein L32; K... 135 3e-32
tpv:TP04_0094 60S ribosomal protein L32; K02912 large subunit ... 135 4e-32
dre:556556 MGC136952, MGC174241, RPL32; zgc:136952; K02912 lar... 134 8e-32
mmu:245405 Gm4987, EG245405; predicted gene 4987 124 9e-29
mmu:621697 Gm6251, EG621697; predicted gene 6251 122 3e-28
cel:T24B8.1 rpl-32; Ribosomal Protein, Large subunit family me... 107 9e-24
sce:YBL092W RPL32; L32; K02912 large subunit ribosomal protein... 105 4e-23
ath:AT5G08420 RNA binding; K06961 ribosomal RNA assembly protein 28.1 7.1
hsa:64215 DNAJC1, DNAJL1, ERdj1, HTJ1, MGC131954, MTJ1; DnaJ (... 27.7 9.2
> tgo:TGME49_067400 ribosomal protein L32, putative ; K02912 large
subunit ribosomal protein L32e
Length=134
Score = 204 bits (519), Expect = 5e-53, Method: Compositional matrix adjust.
Identities = 96/118 (81%), Positives = 106/118 (89%), Gaps = 0/118 (0%)
Query 12 MAPVSTVKRRIVKKRTKRFPRFQSDRWKRLSPSWRKPKGIDCRVRRKFKGTNLMPNIGYG 71
MAPVSTVKR IVKKR K FPRFQSDR+KR+ SWRKPKGIDCRVRRKFKGTN MPNIGYG
Sbjct 1 MAPVSTVKRSIVKKRVKSFPRFQSDRYKRVKSSWRKPKGIDCRVRRKFKGTNTMPNIGYG 60
Query 72 SNKRTRHMLRNGFYKFRVERPQDIDMLLMHNTKFAAEIASGVSSRKRREIVERAEQLN 129
SNK+TRHML NGF+KF V P+DI++LLMHNTKFAAEIA +SSRKRREI+ERA+QLN
Sbjct 61 SNKKTRHMLPNGFFKFLVSSPKDIELLLMHNTKFAAEIAHNISSRKRREILERADQLN 118
> cpv:cgd6_3340 60S ribosomal protein L32 ; K02912 large subunit
ribosomal protein L32e
Length=137
Score = 172 bits (436), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 81/111 (72%), Positives = 92/111 (82%), Gaps = 0/111 (0%)
Query 19 KRRIVKKRTKRFPRFQSDRWKRLSPSWRKPKGIDCRVRRKFKGTNLMPNIGYGSNKRTRH 78
K+ I+KKRTK+F RFQSDR+ R+ P+WRKPKGIDCRVRRKFKG LMP IGYGS+ +TR
Sbjct 11 KKAIIKKRTKKFTRFQSDRFLRVKPNWRKPKGIDCRVRRKFKGNYLMPKIGYGSDAKTRK 70
Query 79 MLRNGFYKFRVERPQDIDMLLMHNTKFAAEIASGVSSRKRREIVERAEQLN 129
ML NG YKF V PQ++ MLLMHN F EIASGVSSRKRREI+ERAEQLN
Sbjct 71 MLPNGLYKFTVSNPQEVHMLLMHNKTFCVEIASGVSSRKRREILERAEQLN 121
> bbo:BBOV_III002830 17.m07268; 60S ribosomal protein L32; K02912
large subunit ribosomal protein L32e
Length=132
Score = 147 bits (372), Expect = 6e-36, Method: Compositional matrix adjust.
Identities = 71/112 (63%), Positives = 88/112 (78%), Gaps = 0/112 (0%)
Query 18 VKRRIVKKRTKRFPRFQSDRWKRLSPSWRKPKGIDCRVRRKFKGTNLMPNIGYGSNKRTR 77
V R +V KRTK+F RFQSDR+KR+ SWRKPKGIDCRVRR+FKGT L P IGYG++KRTR
Sbjct 5 VFRGLVHKRTKKFRRFQSDRFKRVKESWRKPKGIDCRVRRRFKGTVLTPKIGYGTDKRTR 64
Query 78 HMLRNGFYKFRVERPQDIDMLLMHNTKFAAEIASGVSSRKRREIVERAEQLN 129
H L +G+YK V P ++D+LLMHN AEIA VS++KRR IV+RA++L
Sbjct 65 HKLPSGYYKVVVNNPAEVDILLMHNKTHVAEIAHSVSAKKRRVIVDRAKKLG 116
> pfa:PFI0190w 60S ribosomal protein L32, putative; K02912 large
subunit ribosomal protein L32e
Length=131
Score = 139 bits (350), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 60/104 (57%), Positives = 89/104 (85%), Gaps = 0/104 (0%)
Query 26 RTKRFPRFQSDRWKRLSPSWRKPKGIDCRVRRKFKGTNLMPNIGYGSNKRTRHMLRNGFY 85
RTK+F RFQS+R+ R+ P+WRKP+GIDCRVRR++KGTNLMP+IGYGSNK+T+ +L N Y
Sbjct 13 RTKKFTRFQSNRFMRVKPAWRKPRGIDCRVRRRYKGTNLMPSIGYGSNKKTKFLLPNNKY 72
Query 86 KFRVERPQDIDMLLMHNTKFAAEIASGVSSRKRREIVERAEQLN 129
K+ V+ ++++ L+M++TK+ +IA VSS+KR++I+ERA+Q+N
Sbjct 73 KYVVKNVKEMEPLIMNHTKYCVQIAHNVSSKKRKQIIERAKQMN 116
> mmu:19951 Rpl32, AU020185, rpL32-3A; ribosomal protein L32;
K02912 large subunit ribosomal protein L32e
Length=135
Score = 138 bits (348), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 68/111 (61%), Positives = 87/111 (78%), Gaps = 0/111 (0%)
Query 18 VKRRIVKKRTKRFPRFQSDRWKRLSPSWRKPKGIDCRVRRKFKGTNLMPNIGYGSNKRTR 77
VK +IVKKRTK+F R QSDR+ ++ +WRKP+GID RVRR+FKG LMPNIGYGSNK+T+
Sbjct 8 VKPKIVKKRTKKFIRHQSDRYVKIKRNWRKPRGIDNRVRRRFKGQILMPNIGYGSNKKTK 67
Query 78 HMLRNGFYKFRVERPQDIDMLLMHNTKFAAEIASGVSSRKRREIVERAEQL 128
HML +GF KF V +++++LLM N + AEIA VSS+ R+ IVERA QL
Sbjct 68 HMLPSGFRKFLVHNVKELEVLLMCNKSYCAEIAHNVSSKNRKAIVERAAQL 118
> hsa:6161 RPL32; ribosomal protein L32; K02912 large subunit
ribosomal protein L32e
Length=135
Score = 138 bits (348), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 68/111 (61%), Positives = 87/111 (78%), Gaps = 0/111 (0%)
Query 18 VKRRIVKKRTKRFPRFQSDRWKRLSPSWRKPKGIDCRVRRKFKGTNLMPNIGYGSNKRTR 77
VK +IVKKRTK+F R QSDR+ ++ +WRKP+GID RVRR+FKG LMPNIGYGSNK+T+
Sbjct 8 VKPKIVKKRTKKFIRHQSDRYVKIKRNWRKPRGIDNRVRRRFKGQILMPNIGYGSNKKTK 67
Query 78 HMLRNGFYKFRVERPQDIDMLLMHNTKFAAEIASGVSSRKRREIVERAEQL 128
HML +GF KF V +++++LLM N + AEIA VSS+ R+ IVERA QL
Sbjct 68 HMLPSGFRKFLVHNVKELEVLLMCNKSYCAEIAHNVSSKNRKAIVERAAQL 118
> mmu:100504872 60S ribosomal protein L32-like
Length=135
Score = 138 bits (348), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 68/111 (61%), Positives = 87/111 (78%), Gaps = 0/111 (0%)
Query 18 VKRRIVKKRTKRFPRFQSDRWKRLSPSWRKPKGIDCRVRRKFKGTNLMPNIGYGSNKRTR 77
VK +IVKKRTK+F R QSDR+ ++ +WRKP+GID RVRR+FKG LMPNIGYGSNK+T+
Sbjct 8 VKPKIVKKRTKKFIRHQSDRYVKIKRNWRKPRGIDNRVRRRFKGQILMPNIGYGSNKKTK 67
Query 78 HMLRNGFYKFRVERPQDIDMLLMHNTKFAAEIASGVSSRKRREIVERAEQL 128
HML +GF KF V +++++LLM N + AEIA VSS+ R+ IVERA QL
Sbjct 68 HMLPSGFRKFLVHNVKELEVLLMCNKSYCAEIAHNVSSKNRKAIVERAAQL 118
> ath:AT4G18100 60S ribosomal protein L32 (RPL32A); K02912 large
subunit ribosomal protein L32e
Length=133
Score = 138 bits (348), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 65/115 (56%), Positives = 86/115 (74%), Gaps = 0/115 (0%)
Query 15 VSTVKRRIVKKRTKRFPRFQSDRWKRLSPSWRKPKGIDCRVRRKFKGTNLMPNIGYGSNK 74
V + +++VKKR+ +F R QSDR + SWR+PKGID RVRRKFKG LMPN+GYGS+K
Sbjct 3 VPLLTKKVVKKRSAKFIRPQSDRRITVKESWRRPKGIDSRVRRKFKGVTLMPNVGYGSDK 62
Query 75 RTRHMLRNGFYKFRVERPQDIDMLLMHNTKFAAEIASGVSSRKRREIVERAEQLN 129
+TRH L NGF KF V ++++L+MHN + AEIA VS++KR+ IVERA QL+
Sbjct 63 KTRHYLPNGFKKFVVHNTSELELLMMHNRTYCAEIAHNVSTKKRKAIVERASQLD 117
> ath:AT5G46430 60S ribosomal protein L32 (RPL32B)
Length=133
Score = 137 bits (346), Expect = 7e-33, Method: Compositional matrix adjust.
Identities = 64/115 (55%), Positives = 86/115 (74%), Gaps = 0/115 (0%)
Query 15 VSTVKRRIVKKRTKRFPRFQSDRWKRLSPSWRKPKGIDCRVRRKFKGTNLMPNIGYGSNK 74
V + +++VKKR+ +F R QSDR + SWR+PKGID RVRRKFKG LMPN+GYGS+K
Sbjct 3 VPLLTKKVVKKRSAKFIRPQSDRRITVKESWRRPKGIDSRVRRKFKGVTLMPNVGYGSDK 62
Query 75 RTRHMLRNGFYKFRVERPQDIDMLLMHNTKFAAEIASGVSSRKRREIVERAEQLN 129
+TRH L NGF KF V ++++L+MHN + AEIA +S++KR+ IVERA QL+
Sbjct 63 KTRHYLPNGFKKFIVHNTSELELLMMHNRTYCAEIAHNISTKKRKAIVERASQLD 117
> xla:447129 rpl32, MGC85374, MGC86311; ribosomal protein L32;
K02912 large subunit ribosomal protein L32e
Length=135
Score = 135 bits (341), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 66/111 (59%), Positives = 84/111 (75%), Gaps = 0/111 (0%)
Query 18 VKRRIVKKRTKRFPRFQSDRWKRLSPSWRKPKGIDCRVRRKFKGTNLMPNIGYGSNKRTR 77
K +IVKKRTK+F R QSDR+ ++ +WRKP+GID R RR+FKG LMPNIGYGSNK+T+
Sbjct 8 TKPKIVKKRTKKFIRHQSDRYVKIKRNWRKPRGIDNRARRRFKGQILMPNIGYGSNKKTK 67
Query 78 HMLRNGFYKFRVERPQDIDMLLMHNTKFAAEIASGVSSRKRREIVERAEQL 128
HML GF KF V +++++L+M N F AEIA VSS+ R+ IVERA QL
Sbjct 68 HMLPTGFKKFLVHNIKELEVLMMSNKSFCAEIAHNVSSKNRKTIVERAAQL 118
> tpv:TP04_0094 60S ribosomal protein L32; K02912 large subunit
ribosomal protein L32e
Length=132
Score = 135 bits (339), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 64/107 (59%), Positives = 80/107 (74%), Gaps = 0/107 (0%)
Query 22 IVKKRTKRFPRFQSDRWKRLSPSWRKPKGIDCRVRRKFKGTNLMPNIGYGSNKRTRHMLR 81
+V KRTK+F RF SDR+KRL +WRKPKGID RVRR+FK T LMP IGYG+ KRTRH L
Sbjct 9 LVHKRTKKFRRFHSDRFKRLRTTWRKPKGIDNRVRRRFKSTVLMPKIGYGTCKRTRHQLP 68
Query 82 NGFYKFRVERPQDIDMLLMHNTKFAAEIASGVSSRKRREIVERAEQL 128
G YK V P ++D LLMHN AE+A VS++KRR +++RA++L
Sbjct 69 GGVYKVVVSTPAEVDALLMHNKTHVAEVAHSVSAKKRRVLLDRADKL 115
> dre:556556 MGC136952, MGC174241, RPL32; zgc:136952; K02912 large
subunit ribosomal protein L32e
Length=135
Score = 134 bits (337), Expect = 8e-32, Method: Compositional matrix adjust.
Identities = 65/111 (58%), Positives = 85/111 (76%), Gaps = 0/111 (0%)
Query 18 VKRRIVKKRTKRFPRFQSDRWKRLSPSWRKPKGIDCRVRRKFKGTNLMPNIGYGSNKRTR 77
K +IVKKRTK+F R QSDR+ ++ +WRKP+GID RVRR+FKG LMPNIGYGSN++T+
Sbjct 8 TKPKIVKKRTKKFIRHQSDRYVKIRANWRKPRGIDNRVRRRFKGQMLMPNIGYGSNRKTK 67
Query 78 HMLRNGFYKFRVERPQDIDMLLMHNTKFAAEIASGVSSRKRREIVERAEQL 128
HML +GF KF V +++++L+M N AEIA VSS+ R+ IVERA QL
Sbjct 68 HMLPSGFKKFLVHNVKELEVLMMSNKSHCAEIAHNVSSKNRKLIVERAAQL 118
> mmu:245405 Gm4987, EG245405; predicted gene 4987
Length=135
Score = 124 bits (310), Expect = 9e-29, Method: Compositional matrix adjust.
Identities = 64/114 (56%), Positives = 82/114 (71%), Gaps = 0/114 (0%)
Query 18 VKRRIVKKRTKRFPRFQSDRWKRLSPSWRKPKGIDCRVRRKFKGTNLMPNIGYGSNKRTR 77
VK +IVKKRTK+F R QSDR+ ++ +W KP+GI RVRR+FKG LMPNI Y SNK+T+
Sbjct 8 VKPKIVKKRTKKFIRHQSDRYVKIKQNWWKPRGIYNRVRRRFKGEILMPNISYRSNKKTK 67
Query 78 HMLRNGFYKFRVERPQDIDMLLMHNTKFAAEIASGVSSRKRREIVERAEQLNGR 131
HML + F KF V +++++LLM N + AEIA VSS+ R IVERA QL R
Sbjct 68 HMLPSSFCKFLVHDVKELEVLLMCNKSYCAEIAHNVSSKNREAIVERAAQLAIR 121
> mmu:621697 Gm6251, EG621697; predicted gene 6251
Length=133
Score = 122 bits (306), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 63/111 (56%), Positives = 83/111 (74%), Gaps = 2/111 (1%)
Query 18 VKRRIVKKRTKRFPRFQSDRWKRLSPSWRKPKGIDCRVRRKFKGTNLMPNIGYGSNKRTR 77
VK +IVKKRTK+F R QSDR+ ++ +WRKP+GID RVRR+ G LMPNIGYGSNK+T+
Sbjct 8 VKPKIVKKRTKKFIRHQSDRYVKIKRNWRKPRGIDHRVRRR--GQILMPNIGYGSNKKTK 65
Query 78 HMLRNGFYKFRVERPQDIDMLLMHNTKFAAEIASGVSSRKRREIVERAEQL 128
H L + F KF V +++++LLM N + AEIA V+S+ R+ IVERA QL
Sbjct 66 HTLPSSFRKFLVHNVKELEVLLMCNRSYCAEIAHKVTSKNRKAIVERAAQL 116
> cel:T24B8.1 rpl-32; Ribosomal Protein, Large subunit family
member (rpl-32); K02912 large subunit ribosomal protein L32e
Length=134
Score = 107 bits (267), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 56/98 (57%), Positives = 76/98 (77%), Gaps = 0/98 (0%)
Query 32 RFQSDRWKRLSPSWRKPKGIDCRVRRKFKGTNLMPNIGYGSNKRTRHMLRNGFYKFRVER 91
R +SDR++R++PSWRKPKGID RVRR+F+G MP IG+GS++RTR +L NG+ K V+
Sbjct 21 RHESDRYRRVAPSWRKPKGIDNRVRRRFRGMRAMPTIGHGSDRRTRFVLPNGYKKVLVQN 80
Query 92 PQDIDMLLMHNTKFAAEIASGVSSRKRREIVERAEQLN 129
+D+DMLLM + K+ EI GVS++ R+ IVERA QLN
Sbjct 81 VKDLDMLLMQSYKYIGEIGHGVSAKSRKGIVERAAQLN 118
> sce:YBL092W RPL32; L32; K02912 large subunit ribosomal protein
L32e
Length=130
Score = 105 bits (261), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 51/108 (47%), Positives = 72/108 (66%), Gaps = 0/108 (0%)
Query 21 RIVKKRTKRFPRFQSDRWKRLSPSWRKPKGIDCRVRRKFKGTNLMPNIGYGSNKRTRHML 80
+IVKK TK+F R SDR+ R++ +WRK KGID VRR+F+G P IGYGSNK+T+ +
Sbjct 8 KIVKKHTKKFKRHHSDRYHRVAENWRKQKGIDSVVRRRFRGNISQPKIGYGSNKKTKFLS 67
Query 81 RNGFYKFRVERPQDIDMLLMHNTKFAAEIASGVSSRKRREIVERAEQL 128
+G F V +D++ L MH +AAEIA +S++ R I+ RA+ L
Sbjct 68 PSGHKTFLVANVKDLETLTMHTKTYAAEIAHNISAKNRVVILARAKAL 115
> ath:AT5G08420 RNA binding; K06961 ribosomal RNA assembly protein
Length=391
Score = 28.1 bits (61), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 11/35 (31%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 12 MAPVSTVKRRIVKKRTKRFPRFQSDRWKRLSPSWR 46
M PV +K ++KK ++ P ++ W R P++R
Sbjct 213 MHPVYHIKTLMMKKELEKDPALANESWDRFLPTFR 247
> hsa:64215 DNAJC1, DNAJL1, ERdj1, HTJ1, MGC131954, MTJ1; DnaJ
(Hsp40) homolog, subfamily C, member 1; K09521 DnaJ homolog
subfamily C member 1
Length=554
Score = 27.7 bits (60), Expect = 9.2, Method: Composition-based stats.
Identities = 13/54 (24%), Positives = 28/54 (51%), Gaps = 0/54 (0%)
Query 6 QRSISRMAPVSTVKRRIVKKRTKRFPRFQSDRWKRLSPSWRKPKGIDCRVRRKF 59
+R+ S P + ++++++ +++PR SDRW +++ DC R K
Sbjct 488 ERARSAEEPWTQNQQKLLELALQQYPRGSSDRWDKIARCVPSKSKEDCIARYKL 541
Lambda K H
0.323 0.134 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2105161088
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40