bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7249_orf2
Length=167
Score E
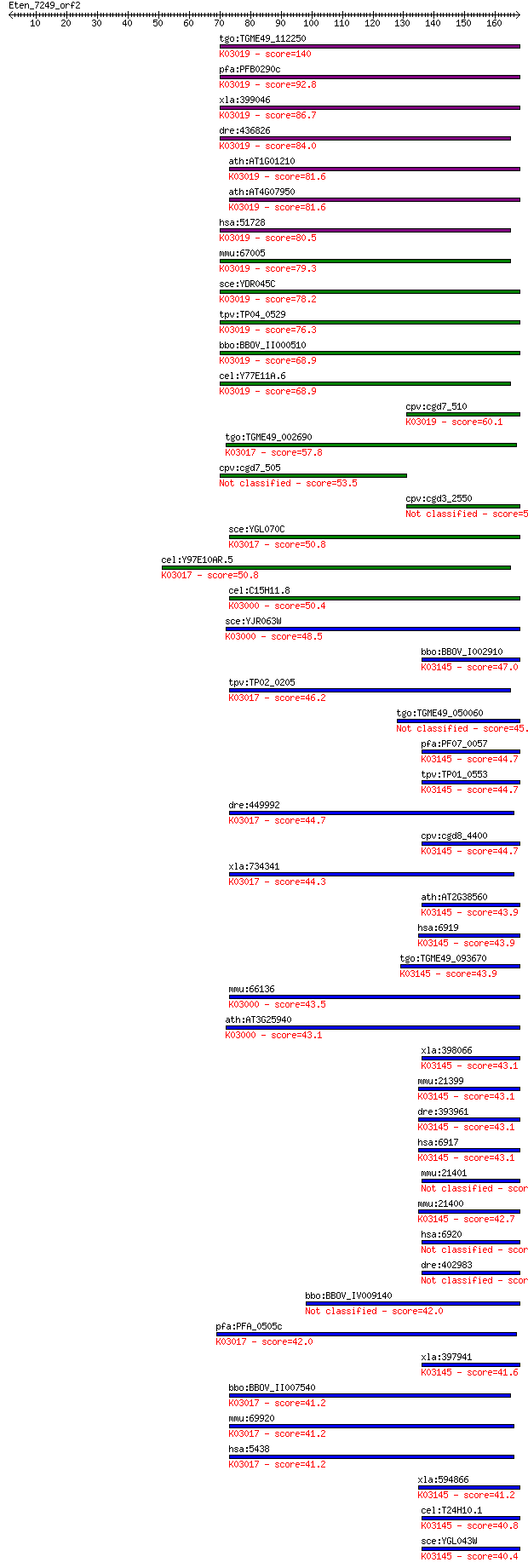
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_112250 DNA-directed RNA polymerase III subunit, put... 140 2e-33
pfa:PFB0290c transcription factor, putative; K03019 DNA-direct... 92.8 5e-19
xla:399046 polr3k, MGC68582; polymerase (RNA) III (DNA directe... 86.7 3e-17
dre:436826 polr3k, zgc:92774; polymerase (RNA) III (DNA direct... 84.0 2e-16
ath:AT1G01210 DNA-directed RNA polymerase III family protein; ... 81.6 1e-15
ath:AT4G07950 DNA-directed RNA polymerase III family protein; ... 81.6 1e-15
hsa:51728 POLR3K, C11, C11-RNP3, RPC10, RPC11, RPC12.5, hRPC11... 80.5 3e-15
mmu:67005 Polr3k, 12.3kDa, 1500004O14Rik, AI196849, AU014893, ... 79.3 5e-15
sce:YDR045C RPC11; C11; K03019 DNA-directed RNA polymerase III... 78.2 1e-14
tpv:TP04_0529 hypothetical protein; K03019 DNA-directed RNA po... 76.3 4e-14
bbo:BBOV_II000510 18.m06026; transcription factor S-II protein... 68.9 6e-12
cel:Y77E11A.6 hypothetical protein; K03019 DNA-directed RNA po... 68.9 7e-12
cpv:cgd7_510 RNA polymerase III subunit C11 ; K03019 DNA-direc... 60.1 3e-09
tgo:TGME49_002690 DNA-directed RNA polymerase II subunit, puta... 57.8 2e-08
cpv:cgd7_505 hypothetical protein 53.5 3e-07
cpv:cgd3_2550 hypothetical protein 51.2 2e-06
sce:YGL070C RPB9, SSU73; B12.6; K03017 DNA-directed RNA polyme... 50.8 2e-06
cel:Y97E10AR.5 rpb-9; RNA Polymerase II (B) subunit family mem... 50.8 2e-06
cel:C15H11.8 hypothetical protein; K03000 DNA-directed RNA pol... 50.4 2e-06
sce:YJR063W RPA12, RRN4; A12.2; K03000 DNA-directed RNA polyme... 48.5 1e-05
bbo:BBOV_I002910 19.m02325; transcription factor S-II (TFIIS)a... 47.0 3e-05
tpv:TP02_0205 DNA-directed RNA polymerase II subunit; K03017 D... 46.2 5e-05
tgo:TGME49_050060 RNA polymerase, putative 45.4 8e-05
pfa:PF07_0057 transcription elongation factor S-II, putative; ... 44.7 1e-04
tpv:TP01_0553 transcription elongation factor SII; K03145 tran... 44.7 2e-04
dre:449992 polr2i, zgc:103515; polymerase (RNA) II (DNA direct... 44.7 2e-04
cpv:cgd8_4400 transcription elongation factor TFIIS ; K03145 t... 44.7 2e-04
xla:734341 polr2i, MGC85139; polymerase (RNA) II (DNA directed... 44.3 2e-04
ath:AT2G38560 TFIIS; TFIIS (TRANSCRIPT ELONGATION FACTOR IIS);... 43.9 2e-04
hsa:6919 TCEA2, TFIIS; transcription elongation factor A (SII)... 43.9 2e-04
tgo:TGME49_093670 transcription elongation factor TFIIS, putat... 43.9 3e-04
mmu:66136 Znrd1, 1110014N07Rik, Rpa12; zinc ribbon domain cont... 43.5 3e-04
ath:AT3G25940 transcription factor S-II (TFIIS) domain-contain... 43.1 4e-04
xla:398066 transcription elongation factor type xTFIIS.l; K031... 43.1 4e-04
mmu:21399 Tcea1, MGC103154, S-II; transcription elongation fac... 43.1 4e-04
dre:393961 tcea2, MGC56325, zgc:56325; transcription elongatio... 43.1 4e-04
hsa:6917 TCEA1, GTF2S, SII, TCEA, TF2S, TFIIS; transcription e... 43.1 4e-04
mmu:21401 Tcea3, S-II, SII-K1; transcription elongation factor... 42.7 5e-04
mmu:21400 Tcea2, AI326274, S-II-T1, SII-T1, Tceat; transcripti... 42.7 6e-04
hsa:6920 TCEA3, TFIIS, TFIIS.H; transcription elongation facto... 42.4 7e-04
dre:402983 tcea3, MGC77732, zgc:77732; transcription elongatio... 42.4 8e-04
bbo:BBOV_IV009140 23.m06447; hypothetical protein 42.0 0.001
pfa:PFA_0505c DNA-directed RNA polymerase 2, putative (EC:2.7.... 42.0 0.001
xla:397941 xTFIIS.oB; TFIIS elongation factor; K03145 transcri... 41.6 0.001
bbo:BBOV_II007540 18.m06627; DNA-directed RNA polymerase 2 sub... 41.2 0.001
mmu:69920 Polr2i, 2810002B19Rik, MGC73656; polymerase (RNA) II... 41.2 0.002
hsa:5438 POLR2I, RPB9, hRPB14.5; polymerase (RNA) II (DNA dire... 41.2 0.002
xla:594866 tcea1, gtf2s, sii, tcea, tf2s; transcription elonga... 41.2 0.002
cel:T24H10.1 hypothetical protein; K03145 transcription elonga... 40.8 0.002
sce:YGL043W DST1, PPR2; Dst1p; K03145 transcription elongation... 40.4 0.003
> tgo:TGME49_112250 DNA-directed RNA polymerase III subunit, putative
; K03019 DNA-directed RNA polymerase III subunit RPC10
Length=106
Score = 140 bits (353), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 64/100 (64%), Positives = 81/100 (81%), Gaps = 3/100 (3%)
Query 70 MALFCPTCHNMLLLRRDAEMEFYCRTCPYIYAIKKKVARKIILSPKKVDEPIGDDLQE-- 127
MALFCPTCHNMLL+R++ M+F+CRTCPY++ IK+K+ RK+ L+PKK DEP+ D+ QE
Sbjct 1 MALFCPTCHNMLLVRQEVTMQFHCRTCPYVFNIKEKLTRKMPLTPKKADEPL-DESQEAA 59
Query 128 TGARIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
GA+ PA CP CSH EA+FY+IQIRS+DEPMT FY C C
Sbjct 60 RGAKAPASCPKCSHTEAYFYEIQIRSADEPMTAFYCCCNC 99
> pfa:PFB0290c transcription factor, putative; K03019 DNA-directed
RNA polymerase III subunit RPC10
Length=106
Score = 92.8 bits (229), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 42/100 (42%), Positives = 60/100 (60%), Gaps = 2/100 (2%)
Query 70 MALFCPTCHNMLLLRRDAEMEFYCRTCPYIYAIKKKVARKIILSPKKVDEPIG--DDLQE 127
MA FCP CHN++L+ + + FYC++C Y Y IK K+ K P+ D +
Sbjct 1 MAFFCPNCHNIVLVHIEKGVYFYCKSCNYKYKIKNKIYNKFDCQQFNKTIPLDAVDINNK 60
Query 128 TGARIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
++ AVCP C++ EA+FY +QIRS+DEP T FY C +C
Sbjct 61 NMSKTQAVCPKCTNDEAYFYTLQIRSADEPSTIFYICVKC 100
> xla:399046 polr3k, MGC68582; polymerase (RNA) III (DNA directed)
polypeptide K, 12.3 kDa; K03019 DNA-directed RNA polymerase
III subunit RPC10
Length=108
Score = 86.7 bits (213), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 45/102 (44%), Positives = 60/102 (58%), Gaps = 4/102 (3%)
Query 70 MALFCPTCHNMLLLRRDAE-MEFYCRTCPYIYAIKKKVARKIILSPKKVDEPIGDDL-QE 127
M LFCPTC N+L++ + F C TCPY++ I +KV + K+VD+ +G E
Sbjct 1 MLLFCPTCGNVLIVEEGQKCYRFACNTCPYVHNINRKVTSRKYPKLKEVDDVLGGSAAWE 60
Query 128 TGARIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYAC--TQC 167
CP C+H A+F QIQ RS+DEPMTTFY C +QC
Sbjct 61 NVDSTAETCPKCAHPRAYFMQIQTRSADEPMTTFYKCCNSQC 102
> dre:436826 polr3k, zgc:92774; polymerase (RNA) III (DNA directed)
polypeptide K; K03019 DNA-directed RNA polymerase III
subunit RPC10
Length=108
Score = 84.0 bits (206), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 43/97 (44%), Positives = 55/97 (56%), Gaps = 2/97 (2%)
Query 70 MALFCPTCHNMLLLRRDAE-MEFYCRTCPYIYAIKKKVARKIILSPKKVDEPIGDDL-QE 127
M LFCPTC N+L++ F C TCPY++ I +KV + K+VD+ +G E
Sbjct 1 MLLFCPTCGNVLIVEEGQRCFRFACNTCPYVHNITRKVNNRKYPKLKEVDDVLGGSAAWE 60
Query 128 TGARIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYAC 164
CP C H A+F QIQ RS+DEPMTTFY C
Sbjct 61 NVDSTAEPCPKCEHPRAYFMQIQTRSADEPMTTFYKC 97
> ath:AT1G01210 DNA-directed RNA polymerase III family protein;
K03019 DNA-directed RNA polymerase III subunit RPC10
Length=106
Score = 81.6 bits (200), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 39/97 (40%), Positives = 56/97 (57%), Gaps = 2/97 (2%)
Query 73 FCPTCHNMLLLRRDAEMEFYCRTCPYIYAIKKKV--ARKIILSPKKVDEPIGDDLQETGA 130
FCPTC N+L F+C TCPY+ I+++V +K +L K ++ + D T A
Sbjct 3 FCPTCGNLLRYEGGGNSRFFCSTCPYVAYIQRQVEIKKKQLLVKKSIEAVVTKDDIPTAA 62
Query 131 RIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
A CP C H +A+F +QIRS+DEP + FY C +C
Sbjct 63 ETEAPCPRCGHDKAYFKSMQIRSADEPESRFYRCLKC 99
> ath:AT4G07950 DNA-directed RNA polymerase III family protein;
K03019 DNA-directed RNA polymerase III subunit RPC10
Length=106
Score = 81.6 bits (200), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 39/97 (40%), Positives = 56/97 (57%), Gaps = 2/97 (2%)
Query 73 FCPTCHNMLLLRRDAEMEFYCRTCPYIYAIKKKV--ARKIILSPKKVDEPIGDDLQETGA 130
FCPTC N+L F+C TCPY+ I+++V +K +L K ++ + D T A
Sbjct 3 FCPTCGNLLRYEGGGSSRFFCSTCPYVANIERRVEIKKKQLLVKKSIEPVVTKDDIPTAA 62
Query 131 RIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
A CP C H +A+F +QIRS+DEP + FY C +C
Sbjct 63 ETEAPCPRCGHDKAYFKSMQIRSADEPESRFYRCLKC 99
> hsa:51728 POLR3K, C11, C11-RNP3, RPC10, RPC11, RPC12.5, hRPC11;
polymerase (RNA) III (DNA directed) polypeptide K, 12.3
kDa; K03019 DNA-directed RNA polymerase III subunit RPC10
Length=108
Score = 80.5 bits (197), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 41/97 (42%), Positives = 53/97 (54%), Gaps = 2/97 (2%)
Query 70 MALFCPTCHNMLLLRRDAE-MEFYCRTCPYIYAIKKKVARKIILSPKKVDEPIGDDL-QE 127
M LFCP C N L++ F C TCPY++ I +KV + K+VD+ +G E
Sbjct 1 MLLFCPGCGNGLIVEEGQRCHRFSCNTCPYVHNITRKVTNRKYPKLKEVDDVLGGAAAWE 60
Query 128 TGARIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYAC 164
CP C H A+F Q+Q RS+DEPMTTFY C
Sbjct 61 NVDSTAESCPKCEHPRAYFMQLQTRSADEPMTTFYKC 97
> mmu:67005 Polr3k, 12.3kDa, 1500004O14Rik, AI196849, AU014893,
C11, RPC10, RPC11; polymerase (RNA) III (DNA directed) polypeptide
K; K03019 DNA-directed RNA polymerase III subunit RPC10
Length=108
Score = 79.3 bits (194), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 41/97 (42%), Positives = 53/97 (54%), Gaps = 2/97 (2%)
Query 70 MALFCPTCHNMLLLRRDAE-MEFYCRTCPYIYAIKKKVARKIILSPKKVDEPIGDDL-QE 127
M LFCP C N L++ F C TCPY++ I +KV + K+VD+ +G E
Sbjct 1 MLLFCPGCGNGLIVEEGQRCHRFACNTCPYVHNITRKVTNRKYPKLKEVDDVLGGAAAWE 60
Query 128 TGARIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYAC 164
CP C H A+F Q+Q RS+DEPMTTFY C
Sbjct 61 NVDSTAEPCPKCEHPRAYFMQLQTRSADEPMTTFYKC 97
> sce:YDR045C RPC11; C11; K03019 DNA-directed RNA polymerase III
subunit RPC10
Length=110
Score = 78.2 bits (191), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 41/104 (39%), Positives = 63/104 (60%), Gaps = 7/104 (6%)
Query 70 MALFCPTCHNMLLLRRDAEMEFY--CRTCPYIYAIKK-KVARKIILSPKKVDEPIG---D 123
M FCP+C+NMLL+ + CR+CPY + I+ ++ + L K+VD+ +G D
Sbjct 1 MLSFCPSCNNMLLITSGDSGVYTLACRSCPYEFPIEGIEIYDRKKLPRKEVDDVLGGGWD 60
Query 124 DLQETGARIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
++ +T + P +C A+F+Q+QIRS+DEPMTTFY C C
Sbjct 61 NVDQTKTQCPNY-DTCGGESAYFFQLQIRSADEPMTTFYKCVNC 103
> tpv:TP04_0529 hypothetical protein; K03019 DNA-directed RNA
polymerase III subunit RPC10
Length=108
Score = 76.3 bits (186), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 42/105 (40%), Positives = 58/105 (55%), Gaps = 10/105 (9%)
Query 70 MALFCPTCHNMLLLRRDAEME--FYCRTCPYIYAIKKKVARKIILSPKKVDEPIGDDLQE 127
M+LFCP CH++L + E + F C C Y + K ++ + + K G L E
Sbjct 1 MSLFCPLCHSILYFEQKPENKSSFSCLRCTYNLPVTKDYHKETVCTNVK---EAGKTLFE 57
Query 128 TGA-----RIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
RIPAVCP+C++ EA+F IQ RS+DEPMT F+ CT C
Sbjct 58 ANEFKHAPRIPAVCPACNNKEAYFMSIQTRSADEPMTQFFVCTAC 102
> bbo:BBOV_II000510 18.m06026; transcription factor S-II protein;
K03019 DNA-directed RNA polymerase III subunit RPC10
Length=109
Score = 68.9 bits (167), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 39/103 (37%), Positives = 55/103 (53%), Gaps = 5/103 (4%)
Query 70 MALFCPTCHNMLLLRRD--AEMEFYCRTCPYIYAIKKKVARKIILSPKKVD---EPIGDD 124
MALFCP CH++L + F C C Y I ++ + + + + + P +
Sbjct 1 MALFCPLCHSVLFFSCNPPQTSTFSCMRCVYELPISRRYHKSTVYTQFEKEIPRSPHSVN 60
Query 125 LQETGARIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
E +I AVCPSC + EA+F IQ RS+DEPMT F+ CT C
Sbjct 61 EFEHAPKIIAVCPSCHNKEAYFMSIQTRSADEPMTQFFVCTAC 103
> cel:Y77E11A.6 hypothetical protein; K03019 DNA-directed RNA
polymerase III subunit RPC10
Length=108
Score = 68.9 bits (167), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 36/97 (37%), Positives = 51/97 (52%), Gaps = 2/97 (2%)
Query 70 MALFCPTCHNMLLLRRDAE-MEFYCRTCPYIYAIKKKVARKIILSPKKVDEPIGDDLQET 128
M FCP C +L + + M F C CPY+ + + V +I K +D+ +G
Sbjct 1 MLTFCPECGCVLQIESGEQCMRFSCPACPYVCPVTQTVTSRIYPKLKDIDDVLGGPGAWA 60
Query 129 GARI-PAVCPSCSHHEAFFYQIQIRSSDEPMTTFYAC 164
A++ CP CSH A+F Q+Q RS+DEP T FY C
Sbjct 61 NAQVTDETCPVCSHGRAYFMQLQTRSADEPSTIFYRC 97
> cpv:cgd7_510 RNA polymerase III subunit C11 ; K03019 DNA-directed
RNA polymerase III subunit RPC10
Length=62
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 23/37 (62%), Positives = 31/37 (83%), Gaps = 0/37 (0%)
Query 131 RIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
++ AVCP CS EA+F+Q+QIRS+DEPMT+FY C +C
Sbjct 19 KLLAVCPKCSFSEAYFFQLQIRSADEPMTSFYTCVKC 55
> tgo:TGME49_002690 DNA-directed RNA polymerase II subunit, putative
(EC:2.7.7.6 3.2.1.3); K03017 DNA-directed RNA polymerase
II subunit RPB9
Length=335
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 38/107 (35%), Positives = 49/107 (45%), Gaps = 12/107 (11%)
Query 72 LFCPTCHNMLLLRRDAE---MEFYCRTCPY-IYAIKKKVARKIILSPK-----KVDEPIG 122
+FCP C+NML R D E ++F CR C Y Y K A+ I+ K D I
Sbjct 7 MFCPECNNMLYPREDKEKRQLQFLCRQCDYSRYPKKTDTAQHIVERTNFNFKSKEDIVIS 66
Query 123 DDLQET---GARIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQ 166
DL + G CP C FYQ+ R +++ MT Y CTQ
Sbjct 67 SDLSKDPTLGRVFDWRCPRCRGVGGVFYQLPERVAEDAMTLVYVCTQ 113
> cpv:cgd7_505 hypothetical protein
Length=108
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 26/62 (41%), Positives = 38/62 (61%), Gaps = 2/62 (3%)
Query 70 MALFCPTCHNMLLLRRDAE-MEFYCRTCPYIYAIKKKVARKIILSPKKVDEPIGDDLQET 128
M FC CHN+LLL+ E M FYC TCPY++ I ++++ PKK++EP D+ E
Sbjct 1 MVQFCIHCHNILLLKEHEERMAFYCPTCPYVFKIVSQISKVTDFVPKKMEEP-SIDMNEI 59
Query 129 GA 130
+
Sbjct 60 AS 61
> cpv:cgd3_2550 hypothetical protein
Length=203
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 20/37 (54%), Positives = 25/37 (67%), Gaps = 0/37 (0%)
Query 131 RIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
+I +CP CSH+EAFF Q Q RS+DE T Y C +C
Sbjct 159 KIQEICPECSHNEAFFTQFQARSADEGTTVMYECCKC 195
> sce:YGL070C RPB9, SSU73; B12.6; K03017 DNA-directed RNA polymerase
II subunit RPB9
Length=122
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 36/104 (34%), Positives = 47/104 (45%), Gaps = 12/104 (11%)
Query 73 FCPTCHNMLLLRRDAEME---FYCRTCPYIY-AIKKKVARKIILSPKKVDEPIGDDLQET 128
FC C+NML R D E F CRTC Y+ A V R +++ + E G +Q+
Sbjct 6 FCRDCNNMLYPREDKENNRLLFECRTCSYVEEAGSPLVYRHELIT--NIGETAGV-VQDI 62
Query 129 GA-----RIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
G+ R CP C E F+Q Q R D M F+ C C
Sbjct 63 GSDPTLPRSDRECPKCHSRENVFFQSQQRRKDTSMVLFFVCLSC 106
> cel:Y97E10AR.5 rpb-9; RNA Polymerase II (B) subunit family member
(rpb-9); K03017 DNA-directed RNA polymerase II subunit
RPB9
Length=167
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 34/129 (26%), Positives = 60/129 (46%), Gaps = 22/129 (17%)
Query 51 AAAPAAAAPAAAAAAGAAAMAL-FCPTCHNMLLLRRDAE---MEFYCRTCPYIYAIKKKV 106
A+PA + +G + + FCP C+NML R D E + + CR C + ++V
Sbjct 35 GASPAPSQNEKPGKSGPGFVGIKFCPECNNMLYPREDKESRVLMYSCRNCEH-----REV 89
Query 107 A-------RKIILSPKKVDEPIGDDLQETGARIPAV----CPSCSHHEAFFYQIQIRSSD 155
A K++ ++ + +GD + + +P CP C +A F+Q Q + ++
Sbjct 90 AANPCIYVNKLVHEIDELTQIVGDIIHD--PTLPKTEEHQCPVCGKSKAVFFQAQTKKAE 147
Query 156 EPMTTFYAC 164
E M +Y C
Sbjct 148 EEMRLYYVC 156
> cel:C15H11.8 hypothetical protein; K03000 DNA-directed RNA polymerase
I subunit RPA12
Length=119
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 47/100 (47%), Gaps = 5/100 (5%)
Query 73 FCPTCHNMLLLRRDAEMEFYCRTCPYIYAIKKKVARKIILSPKKVDEPIGD-----DLQE 127
FC C +L L A C+ C +A+K++V + + K + + D + +
Sbjct 12 FCGYCGAILELPAQAPATVSCKVCSTRWAVKERVDQVVSRVEKIYERTVADTDGIENDES 71
Query 128 TGARIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
A + +C C H +A + +Q RS+DE T FY C +C
Sbjct 72 ADAVVDHICTKCGHSKASYSTMQTRSADEGQTVFYTCLKC 111
> sce:YJR063W RPA12, RRN4; A12.2; K03000 DNA-directed RNA polymerase
I subunit RPA12
Length=125
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 33/114 (28%), Positives = 50/114 (43%), Gaps = 22/114 (19%)
Query 72 LFCPTCHNML-----LLRRDAEMEFYCRTCPYIYAIKKKVARKII-----------LSPK 115
+FC C ++L +L + E C C IY + K++ L K
Sbjct 8 IFCLDCGDLLENPNAVLGSNVE----CSQCKAIYPKSQFSNLKVVTTTADDAFPSSLRAK 63
Query 116 K--VDEPIGDDLQETGARIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
K V + + + GA I CP C + E ++ +Q+RS+DE T FY CT C
Sbjct 64 KSVVKTSLKKNELKDGATIKEKCPQCGNEEMNYHTLQLRSADEGATVFYTCTSC 117
> bbo:BBOV_I002910 19.m02325; transcription factor S-II (TFIIS)and
transcription factor S-II (TFIIS) central domain containing
protein; K03145 transcription elongation factor S-II
Length=302
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 18/32 (56%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 136 CPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
C C E +YQ+Q RSSDEPMTTF C +C
Sbjct 265 CLKCHSMETVYYQLQTRSSDEPMTTFVTCLEC 296
> tpv:TP02_0205 DNA-directed RNA polymerase II subunit; K03017
DNA-directed RNA polymerase II subunit RPB9
Length=167
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 30/110 (27%), Positives = 47/110 (42%), Gaps = 24/110 (21%)
Query 73 FCPTCHNMLLLRRDA---EMEFYCRTCPY-------------IYAIKKKVARK--IILSP 114
FCP C+N+L + D ++ ++CR C + IY +A K I +SP
Sbjct 6 FCPECNNILYAKADILKRQLRYFCRQCDFSRISDSKSSEDNCIYRTDYNIAGKENIFVSP 65
Query 115 KKVDEPIGDDLQETGARIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYAC 164
V +P G C C H +A F+Q+ R +D+ M + C
Sbjct 66 MVVKDPT------LGRTTRWRCLKCGHQKAVFFQLPERVTDDAMMLVFVC 109
> tgo:TGME49_050060 RNA polymerase, putative
Length=348
Score = 45.4 bits (106), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 20/44 (45%), Positives = 25/44 (56%), Gaps = 4/44 (9%)
Query 128 TGARIPAVC----PSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
+GA+ VC +C H EAFF Q RS+DE MT Y C +C
Sbjct 297 SGAKKGVVCKETCEACGHGEAFFSTFQARSADEGMTVMYECVKC 340
> pfa:PF07_0057 transcription elongation factor S-II, putative;
K03145 transcription elongation factor S-II
Length=403
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 16/32 (50%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 136 CPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
C C ++ ++Q+Q RSSDEPMTTF C +C
Sbjct 366 CFKCKGYDTIYHQLQTRSSDEPMTTFVTCLKC 397
> tpv:TP01_0553 transcription elongation factor SII; K03145 transcription
elongation factor S-II
Length=324
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 17/32 (53%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 136 CPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
C C+ +YQ+Q RSSDEPMTTF C C
Sbjct 287 CNKCNSKVTTYYQLQTRSSDEPMTTFVTCLNC 318
> dre:449992 polr2i, zgc:103515; polymerase (RNA) II (DNA directed)
polypeptide I (EC:2.7.7.6); K03017 DNA-directed RNA polymerase
II subunit RPB9
Length=126
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 31/100 (31%), Positives = 44/100 (44%), Gaps = 7/100 (7%)
Query 73 FCPTCHNMLLLRRDAE---MEFYCRTCPYIYAIKKK--VARKIILSPKKVDEPIGDDLQE 127
FC C+NML + D E + + CR C Y KI ++ + I D Q+
Sbjct 17 FCQECNNMLYPKEDKENRILLYACRNCDYQQEADNSCIYVNKITHEVDELTQIIADVAQD 76
Query 128 -TGARIPAV-CPSCSHHEAFFYQIQIRSSDEPMTTFYACT 165
T R CP C H EA F+Q +++ M +Y CT
Sbjct 77 PTLPRTEDHPCPKCGHKEAVFFQSHSMKAEDAMRLYYVCT 116
> cpv:cgd8_4400 transcription elongation factor TFIIS ; K03145
transcription elongation factor S-II
Length=332
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 16/32 (50%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 136 CPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
C C ++ +YQ+Q RS+DEPMTTF C C
Sbjct 295 CGKCKTNKTTYYQMQTRSADEPMTTFVRCLNC 326
> xla:734341 polr2i, MGC85139; polymerase (RNA) II (DNA directed)
polypeptide I, 14.5kDa; K03017 DNA-directed RNA polymerase
II subunit RPB9
Length=125
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 32/100 (32%), Positives = 43/100 (43%), Gaps = 7/100 (7%)
Query 73 FCPTCHNMLLLRRDAE---MEFYCRTCPYIYAIKKK--VARKIILSPKKVDEPIGDDLQE 127
FC C+NML + D E + + CR C Y KI ++ + I D QE
Sbjct 16 FCQECNNMLYPKEDKENRILLYACRNCDYQQEADNSCIYVNKITHEIDELTQIIADVAQE 75
Query 128 -TGARIPAV-CPSCSHHEAFFYQIQIRSSDEPMTTFYACT 165
T R C C H EA F+Q S++ M +Y CT
Sbjct 76 PTLPRTEDHPCSKCGHKEAVFFQSHSAKSEDAMRLYYVCT 115
> ath:AT2G38560 TFIIS; TFIIS (TRANSCRIPT ELONGATION FACTOR IIS);
DNA binding / RNA polymerase II transcription elongation
factor/ transcription factor; K03145 transcription elongation
factor S-II
Length=378
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 136 CPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
C C + +YQ+Q RS+DEPMTT+ C C
Sbjct 340 CGRCGQRKCTYYQMQTRSADEPMTTYVTCVNC 371
> hsa:6919 TCEA2, TFIIS; transcription elongation factor A (SII),
2; K03145 transcription elongation factor S-II
Length=299
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 16/33 (48%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 135 VCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
C C + Q+Q RSSDEPMTTF C +C
Sbjct 260 TCGKCRKKNCTYTQVQTRSSDEPMTTFVVCNEC 292
> tgo:TGME49_093670 transcription elongation factor TFIIS, putative
; K03145 transcription elongation factor S-II
Length=418
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 18/39 (46%), Positives = 24/39 (61%), Gaps = 2/39 (5%)
Query 129 GARIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
G + P C C + ++Q+Q RSSDEPMTTF C +C
Sbjct 376 GGQFP--CFKCRTTKTVYFQMQTRSSDEPMTTFVTCLEC 412
> mmu:66136 Znrd1, 1110014N07Rik, Rpa12; zinc ribbon domain containing,
1; K03000 DNA-directed RNA polymerase I subunit RPA12
Length=123
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 33/105 (31%), Positives = 47/105 (44%), Gaps = 15/105 (14%)
Query 73 FCPTCHNMLLLRRDAEMEFYCRTCPYIYAIK----KKVARKIILS------PKKVDEPIG 122
FCP C ++L L + C C + ++ K V ++ + P VDE G
Sbjct 16 FCPDCGSVLPLP-GIQDTVICSRCGFSIDVRDCEGKVVKTSVVFNKLGATIPLSVDE--G 72
Query 123 DDLQETGARIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
+LQ G I CP C H ++ Q+RS+DE T FY C C
Sbjct 73 PELQ--GPVIDRRCPRCGHEGMAYHTRQMRSADEGQTVFYTCINC 115
> ath:AT3G25940 transcription factor S-II (TFIIS) domain-containing
protein; K03000 DNA-directed RNA polymerase I subunit
RPA12
Length=119
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 32/105 (30%), Positives = 47/105 (44%), Gaps = 11/105 (10%)
Query 72 LFCPTCHNMLLLRRDAEMEFYCRTCPYIYAIKKKVARKI--ILSPKKVDEPIGDDL--QE 127
LFC C ML+L+ E C C K + ++I +S + + +G L ++
Sbjct 10 LFCNLCGTMLVLKSTKYAE--CPHCKTTRNAKDIIDKEIAYTVSAEDIRRELGISLFGEK 67
Query 128 TGA-----RIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
T A +I C C H E + Q RS+DE TT+Y C C
Sbjct 68 TQAEAELPKIKKACEKCQHPELVYTTRQTRSADEGQTTYYTCPNC 112
> xla:398066 transcription elongation factor type xTFIIS.l; K03145
transcription elongation factor S-II
Length=292
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 136 CPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
C C + Q+Q RS+DEPMTTF C +C
Sbjct 254 CGKCKKKNCTYTQVQTRSADEPMTTFVVCNEC 285
> mmu:21399 Tcea1, MGC103154, S-II; transcription elongation factor
A (SII) 1; K03145 transcription elongation factor S-II
Length=300
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 135 VCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
C C + Q+Q RS+DEPMTTF C +C
Sbjct 261 TCGKCKKKNCTYTQVQTRSADEPMTTFVVCNEC 293
> dre:393961 tcea2, MGC56325, zgc:56325; transcription elongation
factor A (SII), 2; K03145 transcription elongation factor
S-II
Length=300
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 16/33 (48%), Positives = 20/33 (60%), Gaps = 0/33 (0%)
Query 135 VCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
VC C + Q+Q RS+DEPMTTF C +C
Sbjct 261 VCGKCKGKNCTYTQVQTRSADEPMTTFVLCNEC 293
> hsa:6917 TCEA1, GTF2S, SII, TCEA, TF2S, TFIIS; transcription
elongation factor A (SII), 1; K03145 transcription elongation
factor S-II
Length=301
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 135 VCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
C C + Q+Q RS+DEPMTTF C +C
Sbjct 262 TCGKCKKKNCTYTQVQTRSADEPMTTFVVCNEC 294
> mmu:21401 Tcea3, S-II, SII-K1; transcription elongation factor
A (SII), 3
Length=347
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 136 CPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
C C + Q+Q RS+DEPMTTF C +C
Sbjct 309 CSKCKKKNCTYNQVQTRSADEPMTTFVLCNEC 340
> mmu:21400 Tcea2, AI326274, S-II-T1, SII-T1, Tceat; transcription
elongation factor A (SII), 2; K03145 transcription elongation
factor S-II
Length=299
Score = 42.7 bits (99), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 135 VCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
C C + Q+Q RSSDEPMTT+ C +C
Sbjct 260 TCNKCRKKNCTYTQVQTRSSDEPMTTYVVCNEC 292
> hsa:6920 TCEA3, TFIIS, TFIIS.H; transcription elongation factor
A (SII), 3
Length=348
Score = 42.4 bits (98), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 136 CPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
C C + Q+Q RS+DEPMTTF C +C
Sbjct 310 CSKCKKKNCTYNQVQTRSADEPMTTFVLCNEC 341
> dre:402983 tcea3, MGC77732, zgc:77732; transcription elongation
factor A (SII), 3
Length=409
Score = 42.4 bits (98), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 136 CPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
C C + Q+Q RS+DEPMTTF C +C
Sbjct 371 CGKCKKKNCTYNQVQTRSADEPMTTFVLCNEC 402
> bbo:BBOV_IV009140 23.m06447; hypothetical protein
Length=256
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 39/85 (45%), Gaps = 15/85 (17%)
Query 98 YIYAIKK-----KVARKIILSPKKVD-EPIGDDL---------QETGARIPAVCPSCSHH 142
YIY ++ + ++K + K D E GDD + + + +C +C
Sbjct 164 YIYGYRQSCEFDRSSKKWWMDSLKDDGERAGDDNTLFMAKNANKSSREVVKQLCDNCGFE 223
Query 143 EAFFYQIQIRSSDEPMTTFYACTQC 167
EA++ Q RS+DE MT Y C +C
Sbjct 224 EAYYSTFQARSADEGMTVMYECKRC 248
> pfa:PFA_0505c DNA-directed RNA polymerase 2, putative (EC:2.7.7.6);
K03017 DNA-directed RNA polymerase II subunit RPB9
Length=249
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 32/114 (28%), Positives = 47/114 (41%), Gaps = 22/114 (19%)
Query 69 AMALFCPTCHNMLLLRRD---AEMEFYCRTCPYIY-----------AIKKKVARK--IIL 112
A FC C+N+L + D ++ + CR+C YI I RK I +
Sbjct 2 AEVRFCEECNNILYAKSDRKSKKLIYVCRSCEYISLKPNENNNIVARINYNYDRKEDIYI 61
Query 113 SPKKVDEPIGDDLQETGARIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQ 166
P+ ++P G VC C ++EA F Q+ + S PM Y CT
Sbjct 62 HPETKNDPA------LGRVRDWVCKQCGNNEAVFLQLAEKISYNPMALIYVCTN 109
> xla:397941 xTFIIS.oB; TFIIS elongation factor; K03145 transcription
elongation factor S-II
Length=303
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 136 CPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
C C + Q+Q RS+DEPMTTF C +C
Sbjct 265 CGKCKKKNCTYTQVQTRSADEPMTTFVFCNEC 296
> bbo:BBOV_II007540 18.m06627; DNA-directed RNA polymerase 2 subunit;
K03017 DNA-directed RNA polymerase II subunit RPB9
Length=163
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 30/110 (27%), Positives = 42/110 (38%), Gaps = 24/110 (21%)
Query 73 FCPTCHNMLLLRRD---AEMEFYCRTCPYIY---------------AIKKKVARKIILSP 114
FCP C+N+L R D ++ F CR C Y + II+ P
Sbjct 6 FCPECNNILYARPDLTRRQLVFICRQCDYSRWADPGSLLDNCINRTTYNYQTREDIIIPP 65
Query 115 KKVDEPIGDDLQETGARIPAVCPSCSHHEAFFYQIQIRSSDEPMTTFYAC 164
+P G CP C H EA F+Q+ R+ D+ M + C
Sbjct 66 LITKDPT------LGRTRQWDCPRCGHTEATFFQLPERAYDDAMVLVFVC 109
> mmu:69920 Polr2i, 2810002B19Rik, MGC73656; polymerase (RNA)
II (DNA directed) polypeptide I (EC:2.7.7.6); K03017 DNA-directed
RNA polymerase II subunit RPB9
Length=125
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 30/100 (30%), Positives = 43/100 (43%), Gaps = 7/100 (7%)
Query 73 FCPTCHNMLLLRRDAE---MEFYCRTCPYIYAIKKK--VARKIILSPKKVDEPIGDDLQE 127
FC C+NML + D E + + CR C Y KI ++ + I D Q+
Sbjct 16 FCQECNNMLYPKEDKENRILLYACRNCDYQQEADNSCIYVNKITHEVDELTQIIADVSQD 75
Query 128 -TGARIPAV-CPSCSHHEAFFYQIQIRSSDEPMTTFYACT 165
T R C C H EA F+Q +++ M +Y CT
Sbjct 76 PTLPRTEDHPCQKCGHKEAVFFQSHSARAEDAMRLYYVCT 115
> hsa:5438 POLR2I, RPB9, hRPB14.5; polymerase (RNA) II (DNA directed)
polypeptide I, 14.5kDa (EC:2.7.7.6); K03017 DNA-directed
RNA polymerase II subunit RPB9
Length=125
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 30/100 (30%), Positives = 43/100 (43%), Gaps = 7/100 (7%)
Query 73 FCPTCHNMLLLRRDAE---MEFYCRTCPYIYAIKKK--VARKIILSPKKVDEPIGDDLQE 127
FC C+NML + D E + + CR C Y KI ++ + I D Q+
Sbjct 16 FCQECNNMLYPKEDKENRILLYACRNCDYQQEADNSCIYVNKITHEVDELTQIIADVSQD 75
Query 128 -TGARIPAV-CPSCSHHEAFFYQIQIRSSDEPMTTFYACT 165
T R C C H EA F+Q +++ M +Y CT
Sbjct 76 PTLPRTEDHPCQKCGHKEAVFFQSHSARAEDAMRLYYVCT 115
> xla:594866 tcea1, gtf2s, sii, tcea, tf2s; transcription elongation
factor A (SII), 1; K03145 transcription elongation factor
S-II
Length=303
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 18/33 (54%), Gaps = 0/33 (0%)
Query 135 VCPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
C C + Q+Q RS+DEPMTTF C C
Sbjct 264 TCGKCKKKNCTYTQVQTRSADEPMTTFVFCNNC 296
> cel:T24H10.1 hypothetical protein; K03145 transcription elongation
factor S-II
Length=308
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 16/32 (50%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 136 CPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
C C + Q+Q RSSDEPMTTF C +C
Sbjct 270 CGKCGKKNCTYTQLQTRSSDEPMTTFVFCLEC 301
> sce:YGL043W DST1, PPR2; Dst1p; K03145 transcription elongation
factor S-II
Length=309
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 136 CPSCSHHEAFFYQIQIRSSDEPMTTFYACTQC 167
C C + +YQ+Q RS+DEP+TTF C C
Sbjct 271 CGKCKEKKVSYYQLQTRSADEPLTTFCTCEAC 302
Lambda K H
0.323 0.132 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4092719652
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40