bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7278_orf1
Length=64
Score E
Sequences producing significant alignments: (Bits) Value
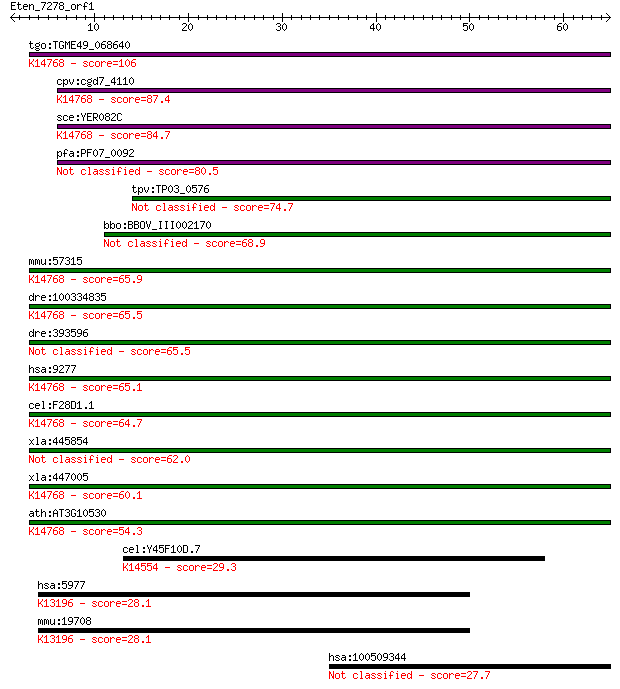
tgo:TGME49_068640 WD domain, G-beta repeat-containing protein ... 106 1e-23
cpv:cgd7_4110 WD40 protein (part of U3 processesome) ; K14768 ... 87.4 1e-17
sce:YER082C UTP7, KRE31; Nucleolar protein, component of the s... 84.7 6e-17
pfa:PF07_0092 BING4 (NUC141) WD-40 repeat protein, putative 80.5 1e-15
tpv:TP03_0576 hypothetical protein 74.7 7e-14
bbo:BBOV_III002170 17.m07209; hypothetical protein 68.9 4e-12
mmu:57315 Wdr46, 2310007I04Rik, Bing4, C78447, C78559; WD repe... 65.9 3e-11
dre:100334835 WD repeat domain 46-like; K14768 U3 small nucleo... 65.5 4e-11
dre:393596 wdr46, Bing4-like, MGC63793, bing4, c6orf11l, zgc:6... 65.5 5e-11
hsa:9277 WDR46, BING4, C6orf11, FP221; WD repeat domain 46; K1... 65.1 5e-11
cel:F28D1.1 hypothetical protein; K14768 U3 small nucleolar RN... 64.7 8e-11
xla:445854 wdr46-b, Bing4, bing4-b; WD repeat domain 46 62.0
xla:447005 wdr46-a, Bing4, MGC82479, bing4-a, wdr46; WD repeat... 60.1 2e-09
ath:AT3G10530 transducin family protein / WD-40 repeat family ... 54.3 1e-07
cel:Y45F10D.7 hypothetical protein; K14554 U3 small nucleolar ... 29.3 3.2
hsa:5977 DPF2, MGC10180, REQ, UBID4, ubi-d4; D4, zinc and doub... 28.1 6.4
mmu:19708 Dpf2, 2210010M07Rik, Req, ubi-d4; D4, zinc and doubl... 28.1 7.2
hsa:100509344 hypothetical protein LOC100509344 27.7 8.5
> tgo:TGME49_068640 WD domain, G-beta repeat-containing protein
; K14768 U3 small nucleolar RNA-associated protein 7
Length=568
Score = 106 bits (265), Expect = 1e-23, Method: Composition-based stats.
Identities = 47/62 (75%), Positives = 52/62 (83%), Gaps = 0/62 (0%)
Query 3 RRPFLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKG 62
R FLPYHYL+VS GEFGEL Y DISTGQIAA+H+T+RGPC M NP+NAVMHLGH KG
Sbjct 230 RMDFLPYHYLLVSVGEFGELVYRDISTGQIAARHKTRRGPCDCMRQNPSNAVMHLGHIKG 289
Query 63 TV 64
TV
Sbjct 290 TV 291
> cpv:cgd7_4110 WD40 protein (part of U3 processesome) ; K14768
U3 small nucleolar RNA-associated protein 7
Length=529
Score = 87.4 bits (215), Expect = 1e-17, Method: Composition-based stats.
Identities = 37/59 (62%), Positives = 45/59 (76%), Gaps = 0/59 (0%)
Query 6 FLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKGTV 64
FLPYH+L+ S GEFGEL Y D+STGQ+AA H+T RGPC +M N N V+HLGH+ G V
Sbjct 192 FLPYHFLLCSIGEFGELRYQDVSTGQVAALHKTGRGPCHIMKHNQFNGVIHLGHSDGVV 250
> sce:YER082C UTP7, KRE31; Nucleolar protein, component of the
small subunit (SSU) processome containing the U3 snoRNA that
is involved in processing of pre-18S rRNA; K14768 U3 small
nucleolar RNA-associated protein 7
Length=554
Score = 84.7 bits (208), Expect = 6e-17, Method: Composition-based stats.
Identities = 38/59 (64%), Positives = 46/59 (77%), Gaps = 0/59 (0%)
Query 6 FLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKGTV 64
FLPYHYL+V+ GE G L Y+D+STGQ+ ++ RTK GP MA NP NAVMHLGH+ GTV
Sbjct 198 FLPYHYLLVTAGETGWLKYHDVSTGQLVSELRTKAGPTMAMAQNPWNAVMHLGHSNGTV 256
> pfa:PF07_0092 BING4 (NUC141) WD-40 repeat protein, putative
Length=602
Score = 80.5 bits (197), Expect = 1e-15, Method: Composition-based stats.
Identities = 33/59 (55%), Positives = 42/59 (71%), Gaps = 0/59 (0%)
Query 6 FLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKGTV 64
FLPYH+L+ S GEFGEL Y DIS G I + +TKRGPC +M N +A+++LGH G V
Sbjct 311 FLPYHFLLTSIGEFGELVYQDISVGNIVTRKKTKRGPCSIMKQNKKDAIIYLGHKNGHV 369
> tpv:TP03_0576 hypothetical protein
Length=515
Score = 74.7 bits (182), Expect = 7e-14, Method: Composition-based stats.
Identities = 33/51 (64%), Positives = 38/51 (74%), Gaps = 0/51 (0%)
Query 14 VSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKGTV 64
V+ GEFGEL Y D+STG+I AKH TK+GPC VM N NAV+HLGH G V
Sbjct 187 VTIGEFGELCYQDVSTGEIVAKHNTKKGPCHVMCQNKDNAVIHLGHKDGLV 237
> bbo:BBOV_III002170 17.m07209; hypothetical protein
Length=525
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 27/54 (50%), Positives = 40/54 (74%), Gaps = 0/54 (0%)
Query 11 YLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKGTV 64
+L+ S GEFGEL + D+S+GQ+ A+++T++GPC +M N N V+HLGH G V
Sbjct 187 WLLTSIGEFGELAWQDVSSGQVVARYKTRKGPCRLMRHNKDNGVVHLGHGNGVV 240
> mmu:57315 Wdr46, 2310007I04Rik, Bing4, C78447, C78559; WD repeat
domain 46; K14768 U3 small nucleolar RNA-associated protein
7
Length=622
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 31/62 (50%), Positives = 40/62 (64%), Gaps = 0/62 (0%)
Query 3 RRPFLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKG 62
R FLP+H+L+ +T E G L Y D+S G+I + G VMA NP NAV+HLGH+ G
Sbjct 279 RLEFLPFHFLLATTSETGFLTYLDVSVGKIVTALNVRAGRLSVMAQNPYNAVIHLGHSNG 338
Query 63 TV 64
TV
Sbjct 339 TV 340
> dre:100334835 WD repeat domain 46-like; K14768 U3 small nucleolar
RNA-associated protein 7
Length=583
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 31/62 (50%), Positives = 38/62 (61%), Gaps = 0/62 (0%)
Query 3 RRPFLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKG 62
R FLPYH+L+ + G L Y D+S G+ A TK G VM NP NA++HLGHT G
Sbjct 250 RMQFLPYHFLLATASATGFLQYLDVSIGKEIAAICTKSGRLNVMTQNPYNAIIHLGHTNG 309
Query 63 TV 64
TV
Sbjct 310 TV 311
> dre:393596 wdr46, Bing4-like, MGC63793, bing4, c6orf11l, zgc:63793;
WD repeat domain 46
Length=583
Score = 65.5 bits (158), Expect = 5e-11, Method: Composition-based stats.
Identities = 31/62 (50%), Positives = 38/62 (61%), Gaps = 0/62 (0%)
Query 3 RRPFLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKG 62
R FLPYH+L+ + G L Y D+S G+ A TK G VM NP NA++HLGHT G
Sbjct 250 RMQFLPYHFLLATASATGFLQYLDVSIGKEIAAICTKSGRLNVMTQNPYNAIIHLGHTSG 309
Query 63 TV 64
TV
Sbjct 310 TV 311
> hsa:9277 WDR46, BING4, C6orf11, FP221; WD repeat domain 46;
K14768 U3 small nucleolar RNA-associated protein 7
Length=556
Score = 65.1 bits (157), Expect = 5e-11, Method: Composition-based stats.
Identities = 30/62 (48%), Positives = 40/62 (64%), Gaps = 0/62 (0%)
Query 3 RRPFLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKG 62
R FLP+H+L+ + E G L Y D+S G+I A + G VM+ NP NAV+HLGH+ G
Sbjct 226 RLEFLPFHFLLATASETGFLTYLDVSVGKIVAALNARAGRLDVMSQNPYNAVIHLGHSNG 285
Query 63 TV 64
TV
Sbjct 286 TV 287
> cel:F28D1.1 hypothetical protein; K14768 U3 small nucleolar
RNA-associated protein 7
Length=580
Score = 64.7 bits (156), Expect = 8e-11, Method: Composition-based stats.
Identities = 30/62 (48%), Positives = 38/62 (61%), Gaps = 0/62 (0%)
Query 3 RRPFLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKG 62
R FLP+H+L+V + LNY D+S G+ A TK G VM NP NA++H GHT G
Sbjct 253 RLEFLPHHFLLVGSSRNSFLNYVDVSVGKQVASFATKSGTLDVMCQNPANAIIHTGHTNG 312
Query 63 TV 64
TV
Sbjct 313 TV 314
> xla:445854 wdr46-b, Bing4, bing4-b; WD repeat domain 46
Length=586
Score = 62.0 bits (149), Expect = 5e-10, Method: Composition-based stats.
Identities = 29/62 (46%), Positives = 37/62 (59%), Gaps = 0/62 (0%)
Query 3 RRPFLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKG 62
R FLPYH+L+ + G L Y D+S G+ A K G VM NP+NA++HLGH G
Sbjct 256 RMEFLPYHFLLATCSSTGFLQYLDVSVGKEIAATCVKSGRLNVMCQNPSNAIIHLGHHNG 315
Query 63 TV 64
TV
Sbjct 316 TV 317
> xla:447005 wdr46-a, Bing4, MGC82479, bing4-a, wdr46; WD repeat
domain 46; K14768 U3 small nucleolar RNA-associated protein
7
Length=586
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 28/62 (45%), Positives = 36/62 (58%), Gaps = 0/62 (0%)
Query 3 RRPFLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKG 62
R FLPYH+L+ + G L Y D+S G+ K G VM NP+NA++HLGH G
Sbjct 256 RMEFLPYHFLLATCSSTGFLQYLDVSVGKEITATCVKSGRLNVMCQNPSNAIIHLGHHNG 315
Query 63 TV 64
TV
Sbjct 316 TV 317
> ath:AT3G10530 transducin family protein / WD-40 repeat family
protein; K14768 U3 small nucleolar RNA-associated protein
7
Length=536
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 27/62 (43%), Positives = 38/62 (61%), Gaps = 0/62 (0%)
Query 3 RRPFLPYHYLMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHLGHTKG 62
R FL H+L+ S G+L+Y D++ G + A RT +G VM NP N+V+ LGH+ G
Sbjct 202 RLRFLKNHFLLASVNMSGQLHYQDVTHGGMVASIRTGKGRTDVMEVNPYNSVVGLGHSGG 261
Query 63 TV 64
TV
Sbjct 262 TV 263
> cel:Y45F10D.7 hypothetical protein; K14554 U3 small nucleolar
RNA-associated protein 21
Length=893
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 13 MVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPNPTNAVMHL 57
++ST G + ++ +ST I AK K G+ AP P+N+++ L
Sbjct 483 LMSTDAQGHILFWSLSTKAINAKMFKKDVRLGICAPCPSNSLVAL 527
> hsa:5977 DPF2, MGC10180, REQ, UBID4, ubi-d4; D4, zinc and double
PHD fingers family 2; K13196 zinc finger protein ubi-d4
Length=391
Score = 28.1 bits (61), Expect = 6.4, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 24/50 (48%), Gaps = 4/50 (8%)
Query 4 RPFLPYHY----LMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPN 49
RP L YHY L GE E + Q + + ++K+GP G+ PN
Sbjct 221 RPGLSYHYAHSHLAEEEGEDKEDSQPPTPVSQRSEEQKSKKGPDGLALPN 270
> mmu:19708 Dpf2, 2210010M07Rik, Req, ubi-d4; D4, zinc and double
PHD fingers family 2; K13196 zinc finger protein ubi-d4
Length=391
Score = 28.1 bits (61), Expect = 7.2, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 24/50 (48%), Gaps = 4/50 (8%)
Query 4 RPFLPYHY----LMVSTGEFGELNYYDISTGQIAAKHRTKRGPCGVMAPN 49
RP L YHY L GE E + Q + + ++K+GP G+ PN
Sbjct 221 RPGLSYHYAHSHLAEEEGEDKEDSRPPTPVSQRSEEQKSKKGPDGLALPN 270
> hsa:100509344 hypothetical protein LOC100509344
Length=543
Score = 27.7 bits (60), Expect = 8.5, Method: Composition-based stats.
Identities = 12/30 (40%), Positives = 17/30 (56%), Gaps = 0/30 (0%)
Query 35 KHRTKRGPCGVMAPNPTNAVMHLGHTKGTV 64
KH + R P ++PNPT++ LG T V
Sbjct 205 KHPSHRPPASTLSPNPTSSTESLGETPHAV 234
Lambda K H
0.321 0.138 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2051595972
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40