bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7310_orf1
Length=123
Score E
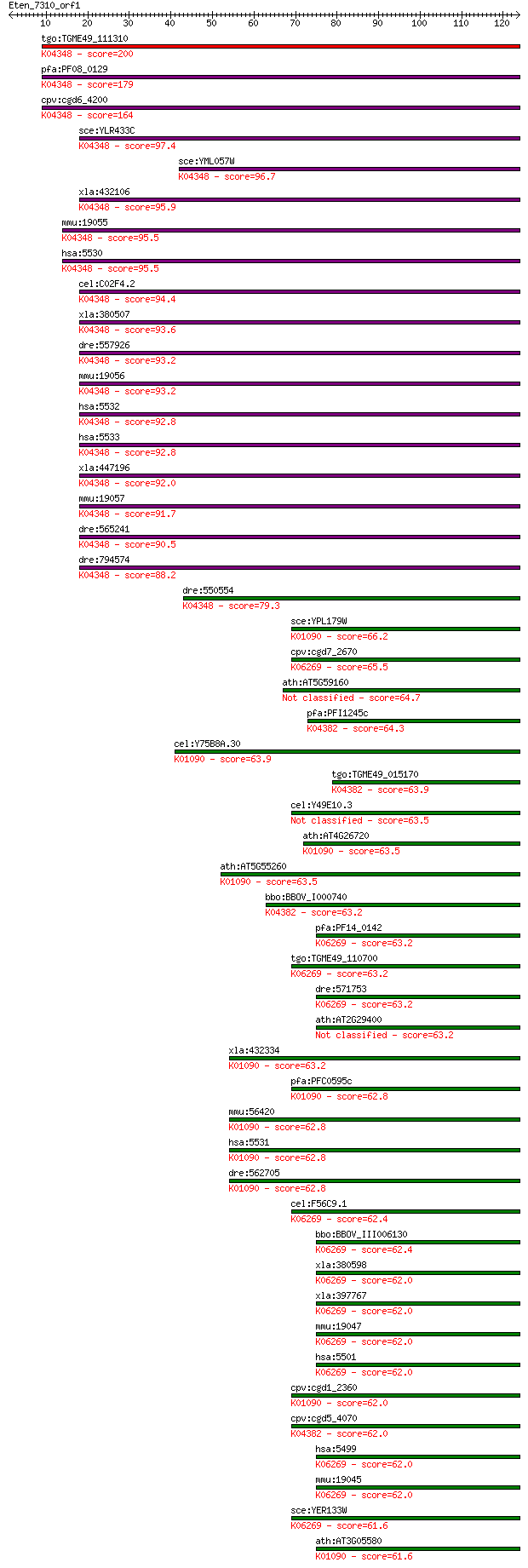
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_111310 serine/threonine protein phosphatase, putati... 200 1e-51
pfa:PF08_0129 serine/threonine protein phosphatase, putative; ... 179 2e-45
cpv:cgd6_4200 protein phosphatase 2B catalytic subunit, calcin... 164 8e-41
sce:YLR433C CNA1, CMP1; Calcineurin A; one isoform (the other ... 97.4 9e-21
sce:YML057W CMP2, CNA2; Calcineurin A; one isoform (the other ... 96.7 1e-20
xla:432106 ppp3cb, MGC81043, calna2, calnb, cna2, pp2bbeta; pr... 95.9 3e-20
mmu:19055 Ppp3ca, 2900074D19Rik, AI841391, AW413465, CN, Caln,... 95.5 3e-20
hsa:5530 PPP3CA, CALN, CALNA, CALNA1, CCN1, CNA1, PPP2B; prote... 95.5 4e-20
cel:C02F4.2 tax-6; abnormal CHEmotaxis family member (tax-6); ... 94.4 8e-20
xla:380507 ppp3ca, MGC52521; protein phosphatase 3 (formerly 2... 93.6 1e-19
dre:557926 ppp3cca, ppp3cc, sb:cb767, si:dkey-21k4.5; protein ... 93.2 2e-19
mmu:19056 Ppp3cb, 1110063J16Rik, Calnb, CnAbeta, Cnab; protein... 93.2 2e-19
hsa:5532 PPP3CB, CALNA2, CALNB, CNA2, PP2Bbeta; protein phosph... 92.8 2e-19
hsa:5533 PPP3CC, CALNA3, CNA3, PP2Bgamma; protein phosphatase ... 92.8 2e-19
xla:447196 ppp3ca, MGC79144, calcineurin, caln, calna, calna1,... 92.0 4e-19
mmu:19057 Ppp3cc, Calnc; protein phosphatase 3, catalytic subu... 91.7 6e-19
dre:565241 ppp3ccb, im:7163784, zgc:158685; protein phosphatas... 90.5 1e-18
dre:794574 ppp3ca, fj46e08, si:dkeyp-79c2.2, wu:fj46e08; prote... 88.2 5e-18
dre:550554 ppp3cb, wu:fq19c08, zgc:110141; protein phosphatase... 79.3 2e-15
sce:YPL179W PPQ1, SAL6; Ppq1p (EC:3.1.3.16); K01090 protein ph... 66.2 2e-11
cpv:cgd7_2670 serine/threonine protein phosphatase ; K06269 pr... 65.5 4e-11
ath:AT5G59160 TOPP2; TOPP2; protein serine/threonine phosphatase 64.7 6e-11
pfa:PFI1245c protein phosphatase-beta; K04382 protein phosphat... 64.3 8e-11
cel:Y75B8A.30 pph-4.1; Protein PHosphatase family member (pph-... 63.9 1e-10
tgo:TGME49_015170 serine/threonine protein phosphatase, putati... 63.9 1e-10
cel:Y49E10.3 pph-4.2; Protein PHosphatase family member (pph-4.2) 63.5 2e-10
ath:AT4G26720 PPX1; PPX1 (PROTEIN PHOSPHATASE X 1); protein se... 63.5 2e-10
ath:AT5G55260 PPX2; PPX2 (PROTEIN PHOSPHATASE X 2); protein se... 63.5 2e-10
bbo:BBOV_I000740 16.m00696; serine/threonine protein phosphata... 63.2 2e-10
pfa:PF14_0142 PP1; serine/threonine protein phosphatase; K0626... 63.2 2e-10
tgo:TGME49_110700 serine/threonine protein phosphatase, putati... 63.2 2e-10
dre:571753 ppp1cc, fe06g11, fi22e08, wu:fe06g11, wu:fi22e08; p... 63.2 2e-10
ath:AT2G29400 TOPP1; TOPP1 (TYPE ONE PROTEIN PHOSPHATASE 1); p... 63.2 2e-10
xla:432334 ppp4c, MGC78774; protein phosphatase 4, catalytic s... 63.2 2e-10
pfa:PFC0595c serine/threonine protein phosphatase, putative; K... 62.8 2e-10
mmu:56420 Ppp4c, 1110002D08Rik, AU016079, Ppx; protein phospha... 62.8 2e-10
hsa:5531 PPP4C, PP4, PP4C, PPH3, PPP4, PPX; protein phosphatas... 62.8 2e-10
dre:562705 ppp4cb, PP4C-B, zgc:172126; protein phosphatase 4 (... 62.8 3e-10
cel:F56C9.1 gsp-2; yeast Glc Seven-like Phosphatases family me... 62.4 3e-10
bbo:BBOV_III006130 17.m07543; Ser/Thr protein phosphatase fami... 62.4 3e-10
xla:380598 ppp1cc, MGC64327, PP-1G-B, ppp1cc-B, xPP1-zeta; pro... 62.0 4e-10
xla:397767 ppp1cc, MGC85040, ppp1cc-a, ppp1g; protein phosphat... 62.0 4e-10
mmu:19047 Ppp1cc, PP1, dis2m1; protein phosphatase 1, catalyti... 62.0 4e-10
hsa:5501 PPP1CC, PP1gamma, PPP1G; protein phosphatase 1, catal... 62.0 4e-10
cpv:cgd1_2360 protein phosphatase 4 (formerly X), catalytic su... 62.0 4e-10
cpv:cgd5_4070 hypothetical protein ; K04382 protein phosphatas... 62.0 4e-10
hsa:5499 PPP1CA, MGC15877, MGC1674, PP-1A, PP1alpha, PPP1A; pr... 62.0 4e-10
mmu:19045 Ppp1ca, Ppp1c, dism2; protein phosphatase 1, catalyt... 62.0 5e-10
sce:YER133W GLC7, CID1, DIS2; Glc7p (EC:3.1.3.16); K06269 prot... 61.6 5e-10
ath:AT3G05580 serine/threonine protein phosphatase, putative; ... 61.6 6e-10
> tgo:TGME49_111310 serine/threonine protein phosphatase, putative
(EC:3.1.3.16); K04348 protein phosphatase 3, catalytic
subunit [EC:3.1.3.16]
Length=501
Score = 200 bits (508), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 93/115 (80%), Positives = 105/115 (91%), Gaps = 0/115 (0%)
Query 9 MDPLPDPKHDRVVSSVQPPPAKPLSSSFLFPEGPDAPPNWRELRSHLQREGRVYKEDCLD 68
M+PL DP HDRVV+SVQPPPA+PL S L+P GP PPNW+EL+SHLQREGR+ KEDCL+
Sbjct 1 MEPLADPLHDRVVTSVQPPPAQPLKHSLLYPSGPANPPNWQELKSHLQREGRLSKEDCLE 60
Query 69 IIRRVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
+I+ VTEITSNEPNLLRL+DPITVVGDIHGQ+YDLLKLLDVGGDPDTTQYLFLGD
Sbjct 61 LIKNVTEITSNEPNLLRLNDPITVVGDIHGQFYDLLKLLDVGGDPDTTQYLFLGD 115
> pfa:PF08_0129 serine/threonine protein phosphatase, putative;
K04348 protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=604
Score = 179 bits (454), Expect = 2e-45, Method: Compositional matrix adjust.
Identities = 81/115 (70%), Positives = 99/115 (86%), Gaps = 0/115 (0%)
Query 9 MDPLPDPKHDRVVSSVQPPPAKPLSSSFLFPEGPDAPPNWRELRSHLQREGRVYKEDCLD 68
M+PLPDPK+DR V V+PPPAKPLS L+P G + PP+++ LR HL++EGR+ KEDCLD
Sbjct 1 MEPLPDPKNDRQVKDVEPPPAKPLSLELLYPNGTEEPPDYKALRDHLKKEGRIRKEDCLD 60
Query 69 IIRRVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
II++V +I SNEPNLLRL DPIT+VGDIHGQYYDLLKLL+VGG+PD TQ+LFLGD
Sbjct 61 IIKKVIDIVSNEPNLLRLKDPITIVGDIHGQYYDLLKLLEVGGNPDHTQFLFLGD 115
> cpv:cgd6_4200 protein phosphatase 2B catalytic subunit, calcineurin
like phosphatse superfamily ; K04348 protein phosphatase
3, catalytic subunit [EC:3.1.3.16]
Length=504
Score = 164 bits (414), Expect = 8e-41, Method: Compositional matrix adjust.
Identities = 78/115 (67%), Positives = 91/115 (79%), Gaps = 0/115 (0%)
Query 9 MDPLPDPKHDRVVSSVQPPPAKPLSSSFLFPEGPDAPPNWRELRSHLQREGRVYKEDCLD 68
M PL DP +DRVV PPP P++ LFP G + P+WR L+SHL REGRV K+ L+
Sbjct 1 MQPLKDPLNDRVVKESDPPPPLPITEELLFPNGIEGNPDWRILKSHLLREGRVRKDHLLE 60
Query 69 IIRRVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
I+RR +EIT+NE NLLRL DPITVVGDIHGQYYDL+KLLDVGGDP+TTQYLFLGD
Sbjct 61 IVRRTSEITANESNLLRLRDPITVVGDIHGQYYDLVKLLDVGGDPETTQYLFLGD 115
> sce:YLR433C CNA1, CMP1; Calcineurin A; one isoform (the other
is CMP2) of the catalytic subunit of calcineurin, a Ca++/calmodulin-regulated
protein phosphatase which regulates Crz1p
(a stress-response transcription factor), the other calcineurin
subunit is CNB1 (EC:3.1.3.16); K04348 protein phosphatase
3, catalytic subunit [EC:3.1.3.16]
Length=553
Score = 97.4 bits (241), Expect = 9e-21, Method: Composition-based stats.
Identities = 53/106 (50%), Positives = 63/106 (59%), Gaps = 1/106 (0%)
Query 18 DRVVSSVQPPPAKPLSSSFLFPEGPDAPPNWRELRSHLQREGRVYKEDCLDIIRRVTEIT 77
DR ++SV K S +F + PN LR H EGR+ KE + I+ T
Sbjct 43 DRPIASVPAITGKIPSDEEVF-DSKTGLPNHSFLREHFFHEGRLSKEQAIKILNMSTVAL 101
Query 78 SNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
S EPNLL+L PIT+ GDIHGQYYDLLKL +VGGDP YLFLGD
Sbjct 102 SKEPNLLKLKAPITICGDIHGQYYDLLKLFEVGGDPAEIDYLFLGD 147
> sce:YML057W CMP2, CNA2; Calcineurin A; one isoform (the other
is CNA1) of the catalytic subunit of calcineurin, a Ca++/calmodulin-regulated
protein phosphatase which regulates Crz1p
(a stress-response transcription factor), the other calcineurin
subunit is CNB1 (EC:3.1.3.16); K04348 protein phosphatase
3, catalytic subunit [EC:3.1.3.16]
Length=604
Score = 96.7 bits (239), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 45/82 (54%), Positives = 55/82 (67%), Gaps = 0/82 (0%)
Query 42 PDAPPNWRELRSHLQREGRVYKEDCLDIIRRVTEITSNEPNLLRLSDPITVVGDIHGQYY 101
P+ P LR H +REG++ I+ TE+ S EPNL+ + PITV GDIHGQY+
Sbjct 91 PNGIPRHEFLRDHFKREGKLSAAQAARIVTLATELFSKEPNLISVPAPITVCGDIHGQYF 150
Query 102 DLLKLLDVGGDPDTTQYLFLGD 123
DLLKL +VGGDP TT YLFLGD
Sbjct 151 DLLKLFEVGGDPATTSYLFLGD 172
> xla:432106 ppp3cb, MGC81043, calna2, calnb, cna2, pp2bbeta;
protein phosphatase 3, catalytic subunit, beta isozyme; K04348
protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=507
Score = 95.9 bits (237), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 49/106 (46%), Positives = 67/106 (63%), Gaps = 3/106 (2%)
Query 18 DRVVSSVQPPPAKPLSSSFLFPEGPDAPPNWRELRSHLQREGRVYKEDCLDIIRRVTEIT 77
+R V +V PP + L+ + +F EG P L++HL +EGR+ +E L IIR I
Sbjct 14 ERPVRAVPFPPTRRLTMNEVFVEGR---PQLETLKNHLIKEGRLEEEVALRIIREGAAIL 70
Query 78 SNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
E +L + PITV GDIHGQ++DL+KL +VGG P T+YLFLGD
Sbjct 71 RQEKTMLEVEAPITVCGDIHGQFFDLMKLFEVGGSPQNTRYLFLGD 116
> mmu:19055 Ppp3ca, 2900074D19Rik, AI841391, AW413465, CN, Caln,
Calna, CnA, MGC106804; protein phosphatase 3, catalytic subunit,
alpha isoform (EC:3.1.3.16); K04348 protein phosphatase
3, catalytic subunit [EC:3.1.3.16]
Length=521
Score = 95.5 bits (236), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 52/113 (46%), Positives = 69/113 (61%), Gaps = 5/113 (4%)
Query 14 DPK---HDRVVSSVQPPPAKPLSSSFLFPEGPDAPPNWRELRSHLQREGRVYKEDCLDII 70
DPK DRVV +V PP+ L++ +F D P L++HL +EGR+ + L II
Sbjct 8 DPKLSTTDRVVKAVPFPPSHRLTAKEVFDN--DGKPRVDILKAHLMKEGRLEESVALRII 65
Query 71 RRVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
I E NLL + P+TV GDIHGQ++DL+KL +VGG P T+YLFLGD
Sbjct 66 TEGASILRQEKNLLDIDAPVTVCGDIHGQFFDLMKLFEVGGSPANTRYLFLGD 118
> hsa:5530 PPP3CA, CALN, CALNA, CALNA1, CCN1, CNA1, PPP2B; protein
phosphatase 3, catalytic subunit, alpha isozyme (EC:3.1.3.16);
K04348 protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=521
Score = 95.5 bits (236), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 52/113 (46%), Positives = 69/113 (61%), Gaps = 5/113 (4%)
Query 14 DPK---HDRVVSSVQPPPAKPLSSSFLFPEGPDAPPNWRELRSHLQREGRVYKEDCLDII 70
DPK DRVV +V PP+ L++ +F D P L++HL +EGR+ + L II
Sbjct 8 DPKLSTTDRVVKAVPFPPSHRLTAKEVFDN--DGKPRVDILKAHLMKEGRLEESVALRII 65
Query 71 RRVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
I E NLL + P+TV GDIHGQ++DL+KL +VGG P T+YLFLGD
Sbjct 66 TEGASILRQEKNLLDIDAPVTVCGDIHGQFFDLMKLFEVGGSPANTRYLFLGD 118
> cel:C02F4.2 tax-6; abnormal CHEmotaxis family member (tax-6);
K04348 protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=535
Score = 94.4 bits (233), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 44/106 (41%), Positives = 67/106 (63%), Gaps = 1/106 (0%)
Query 18 DRVVSSVQPPPAKPLSSSFLFPEGPDAPPNWRELRSHLQREGRVYKEDCLDIIRRVTEIT 77
+R V +VQ P ++ L+ ++ + P LR H +EGR+ +E + +I+ + +
Sbjct 39 ERFVKTVQFPVSERLTVDQVY-DRRTGKPRHEVLRDHFIKEGRIEEEAAIRVIQECSSLF 97
Query 78 SNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
NE +L + P+TV GDIHGQ+YDL+KL +VGG P TT+YLFLGD
Sbjct 98 RNEKTMLEIEAPVTVCGDIHGQFYDLMKLFEVGGSPATTKYLFLGD 143
> xla:380507 ppp3ca, MGC52521; protein phosphatase 3 (formerly
2B), catalytic subunit, alpha isoform (calcineurin A alpha)
(EC:3.1.3.16); K04348 protein phosphatase 3, catalytic subunit
[EC:3.1.3.16]
Length=518
Score = 93.6 bits (231), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 48/106 (45%), Positives = 64/106 (60%), Gaps = 2/106 (1%)
Query 18 DRVVSSVQPPPAKPLSSSFLFPEGPDAPPNWRELRSHLQREGRVYKEDCLDIIRRVTEIT 77
DR V +V PP L++ +F D P L++HL +EGR+ + L II I
Sbjct 15 DRAVKAVPFPPNHRLTAKEIFDN--DGKPRVDVLKAHLMKEGRLEESVALRIISEGASIL 72
Query 78 SNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
E NLL + P+TV GDIHGQ++DL+KL +VGG P T+YLFLGD
Sbjct 73 RQEKNLLDIDAPVTVCGDIHGQFFDLMKLFEVGGSPANTRYLFLGD 118
> dre:557926 ppp3cca, ppp3cc, sb:cb767, si:dkey-21k4.5; protein
phosphatase 3, catalytic subunit, gamma isozyme, a (EC:3.1.3.16);
K04348 protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=499
Score = 93.2 bits (230), Expect = 2e-19, Method: Composition-based stats.
Identities = 47/106 (44%), Positives = 68/106 (64%), Gaps = 3/106 (2%)
Query 18 DRVVSSVQPPPAKPLSSSFLFPEGPDAPPNWRELRSHLQREGRVYKEDCLDIIRRVTEIT 77
+R++ +V PP + L+ L+ +G PN L++HL +EGRV ++ L II I
Sbjct 14 ERIIKTVPLPPHQRLTIKDLYVDGK---PNTELLKNHLIKEGRVEEDAALRIINDGANIL 70
Query 78 SNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
E +L + PITV GD+HGQ++DL+KL +VGG PD T+YLFLGD
Sbjct 71 RQEKCMLEVEAPITVCGDVHGQFFDLMKLFEVGGSPDNTRYLFLGD 116
> mmu:19056 Ppp3cb, 1110063J16Rik, Calnb, CnAbeta, Cnab; protein
phosphatase 3, catalytic subunit, beta isoform (EC:3.1.3.16);
K04348 protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=525
Score = 93.2 bits (230), Expect = 2e-19, Method: Composition-based stats.
Identities = 50/106 (47%), Positives = 65/106 (61%), Gaps = 2/106 (1%)
Query 18 DRVVSSVQPPPAKPLSSSFLFPEGPDAPPNWRELRSHLQREGRVYKEDCLDIIRRVTEIT 77
DRVV +V PP L+S +F D P L++HL +EGRV +E L II I
Sbjct 24 DRVVKAVPFPPTHRLTSEEVFDM--DGIPRVDVLKNHLVKEGRVDEEIALRIINEGAAIL 81
Query 78 SNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
E ++ + PITV GDIHGQ++DL+KL +VGG P T+YLFLGD
Sbjct 82 RREKTMIEVEAPITVCGDIHGQFFDLMKLFEVGGSPANTRYLFLGD 127
> hsa:5532 PPP3CB, CALNA2, CALNB, CNA2, PP2Bbeta; protein phosphatase
3, catalytic subunit, beta isozyme (EC:3.1.3.16); K04348
protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=525
Score = 92.8 bits (229), Expect = 2e-19, Method: Composition-based stats.
Identities = 50/106 (47%), Positives = 65/106 (61%), Gaps = 2/106 (1%)
Query 18 DRVVSSVQPPPAKPLSSSFLFPEGPDAPPNWRELRSHLQREGRVYKEDCLDIIRRVTEIT 77
DRVV +V PP L+S +F D P L++HL +EGRV +E L II I
Sbjct 24 DRVVKAVPFPPTHRLTSEEVF--DLDGIPRVDVLKNHLVKEGRVDEEIALRIINEGAAIL 81
Query 78 SNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
E ++ + PITV GDIHGQ++DL+KL +VGG P T+YLFLGD
Sbjct 82 RREKTMIEVEAPITVCGDIHGQFFDLMKLFEVGGSPANTRYLFLGD 127
> hsa:5533 PPP3CC, CALNA3, CNA3, PP2Bgamma; protein phosphatase
3, catalytic subunit, gamma isozyme (EC:3.1.3.16); K04348
protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=512
Score = 92.8 bits (229), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 47/106 (44%), Positives = 65/106 (61%), Gaps = 3/106 (2%)
Query 18 DRVVSSVQPPPAKPLSSSFLFPEGPDAPPNWRELRSHLQREGRVYKEDCLDIIRRVTEIT 77
DRV+ +V PP + L+ +F G P L++HL +EGR+ +E L II I
Sbjct 12 DRVIKAVPFPPTQRLTFKEVFENGK---PKVDVLKNHLVKEGRLEEEVALKIINDGAAIL 68
Query 78 SNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
E ++ + PITV GDIHGQ++DL+KL +VGG P T+YLFLGD
Sbjct 69 RQEKTMIEVDAPITVCGDIHGQFFDLMKLFEVGGSPSNTRYLFLGD 114
> xla:447196 ppp3ca, MGC79144, calcineurin, caln, calna, calna1,
ccn1, cna1, ppp2b; protein phosphatase 3, catalytic subunit,
alpha isozyme; K04348 protein phosphatase 3, catalytic subunit
[EC:3.1.3.16]
Length=511
Score = 92.0 bits (227), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 47/106 (44%), Positives = 64/106 (60%), Gaps = 2/106 (1%)
Query 18 DRVVSSVQPPPAKPLSSSFLFPEGPDAPPNWRELRSHLQREGRVYKEDCLDIIRRVTEIT 77
DR V +V PP+ L++ +F D P L+ HL +EGR+ + L +I I
Sbjct 15 DRAVKAVPFPPSHRLTAKEVFDN--DGKPRVDILKGHLMKEGRLEESVALRLITEGASIL 72
Query 78 SNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
E NLL + P+TV GDIHGQ++DL+KL +VGG P T+YLFLGD
Sbjct 73 RQEKNLLDIDAPVTVCGDIHGQFFDLMKLFEVGGSPANTRYLFLGD 118
> mmu:19057 Ppp3cc, Calnc; protein phosphatase 3, catalytic subunit,
gamma isoform (EC:3.1.3.16); K04348 protein phosphatase
3, catalytic subunit [EC:3.1.3.16]
Length=513
Score = 91.7 bits (226), Expect = 6e-19, Method: Composition-based stats.
Identities = 46/106 (43%), Positives = 65/106 (61%), Gaps = 3/106 (2%)
Query 18 DRVVSSVQPPPAKPLSSSFLFPEGPDAPPNWRELRSHLQREGRVYKEDCLDIIRRVTEIT 77
+RV+ +V PP + L+ +F G P L++HL +EGRV +E L II I
Sbjct 12 ERVIKAVPFPPTRRLTLKEVFENGK---PKMDLLKNHLVKEGRVEEEVALKIINDGAAIL 68
Query 78 SNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
E ++ + PITV GD+HGQ++DL+KL +VGG P T+YLFLGD
Sbjct 69 KQEKTMIEVEAPITVCGDVHGQFFDLMKLFEVGGSPSNTRYLFLGD 114
> dre:565241 ppp3ccb, im:7163784, zgc:158685; protein phosphatase
3, catalytic subunit, gamma isozyme, b (EC:3.1.3.16); K04348
protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=506
Score = 90.5 bits (223), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 46/106 (43%), Positives = 66/106 (62%), Gaps = 3/106 (2%)
Query 18 DRVVSSVQPPPAKPLSSSFLFPEGPDAPPNWRELRSHLQREGRVYKEDCLDIIRRVTEIT 77
+R+V +V PP L+ L+ +G P L+SHL +EGR+ ++ L II I
Sbjct 13 ERLVKAVPYPPTTRLTMKELYEDGK---PKVELLKSHLVKEGRLEEDVALRIINDGANIL 69
Query 78 SNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
+E +L + PITV GD+HGQ++DL+KL +VGG P T+YLFLGD
Sbjct 70 RHEKCMLEVEAPITVCGDVHGQFFDLMKLFEVGGSPHNTRYLFLGD 115
> dre:794574 ppp3ca, fj46e08, si:dkeyp-79c2.2, wu:fj46e08; protein
phosphatase 3, catalytic subunit, alpha isozyme; K04348
protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=521
Score = 88.2 bits (217), Expect = 5e-18, Method: Composition-based stats.
Identities = 48/106 (45%), Positives = 64/106 (60%), Gaps = 2/106 (1%)
Query 18 DRVVSSVQPPPAKPLSSSFLFPEGPDAPPNWRELRSHLQREGRVYKEDCLDIIRRVTEIT 77
DRVV SV P + L+ +F + P L++HL +EGRV + L II I
Sbjct 14 DRVVKSVPFPLSHRLTMKEVF--DCEGKPRVDLLKAHLTKEGRVEETVALRIINEGASIL 71
Query 78 SNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
E +L + P+TV GDIHGQ++DL+KL +VGG P TT+YLFLGD
Sbjct 72 RQEKTMLDIEAPVTVCGDIHGQFFDLMKLFEVGGSPATTRYLFLGD 117
> dre:550554 ppp3cb, wu:fq19c08, zgc:110141; protein phosphatase
3, catalytic subunit, beta isozyme (EC:3.1.3.16); K04348
protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=511
Score = 79.3 bits (194), Expect = 2e-15, Method: Composition-based stats.
Identities = 37/81 (45%), Positives = 51/81 (62%), Gaps = 0/81 (0%)
Query 43 DAPPNWRELRSHLQREGRVYKEDCLDIIRRVTEITSNEPNLLRLSDPITVVGDIHGQYYD 102
+ P L++HL +EGRV + L I+ I E +L + P+TV GDIHGQ++D
Sbjct 27 EGKPRVDLLKAHLTKEGRVEEPVALRIVNEGAAILRQEKTMLDIEAPVTVCGDIHGQFFD 86
Query 103 LLKLLDVGGDPDTTQYLFLGD 123
L+ L +VGG P TT+YLFLGD
Sbjct 87 LMNLFEVGGSPATTRYLFLGD 107
> sce:YPL179W PPQ1, SAL6; Ppq1p (EC:3.1.3.16); K01090 protein
phosphatase [EC:3.1.3.16]
Length=549
Score = 66.2 bits (160), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 31/55 (56%), Positives = 37/55 (67%), Gaps = 0/55 (0%)
Query 69 IIRRVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
I EI N+P LLRL PI VVGD+HGQ+ DLL++L + G P T YLFLGD
Sbjct 275 ICYHAREIFLNQPTLLRLQAPIKVVGDVHGQFNDLLRILKLSGVPSDTNYLFLGD 329
> cpv:cgd7_2670 serine/threonine protein phosphatase ; K06269
protein phosphatase 1, catalytic subunit [EC:3.1.3.16]
Length=320
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 31/55 (56%), Positives = 37/55 (67%), Gaps = 0/55 (0%)
Query 69 IIRRVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
I + EI ++P LL L PI + GDIHGQYYDLL+L + GG P T YLFLGD
Sbjct 52 ICIKSREIFMSQPTLLELDAPIKICGDIHGQYYDLLRLFEYGGFPPDTNYLFLGD 106
> ath:AT5G59160 TOPP2; TOPP2; protein serine/threonine phosphatase
Length=312
Score = 64.7 bits (156), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 33/57 (57%), Positives = 38/57 (66%), Gaps = 2/57 (3%)
Query 67 LDIIRRVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
L I+ R EI +PNLL L PI + GDIHGQY DLL+L + GG P T YLFLGD
Sbjct 44 LCIVSR--EIFLQQPNLLELEAPIKICGDIHGQYSDLLRLFEYGGFPPTANYLFLGD 98
> pfa:PFI1245c protein phosphatase-beta; K04382 protein phosphatase
2 (formerly 2A), catalytic subunit [EC:3.1.3.16]
Length=466
Score = 64.3 bits (155), Expect = 8e-11, Method: Composition-based stats.
Identities = 27/51 (52%), Positives = 37/51 (72%), Gaps = 0/51 (0%)
Query 73 VTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
+ +I NE N +R++ P+TV GDIHGQ++DLL+L +GG P YLFLGD
Sbjct 186 LIDILKNEENCVRINVPVTVAGDIHGQFFDLLELFHIGGLPPDVNYLFLGD 236
> cel:Y75B8A.30 pph-4.1; Protein PHosphatase family member (pph-4.1);
K01090 protein phosphatase [EC:3.1.3.16]
Length=333
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 32/86 (37%), Positives = 49/86 (56%), Gaps = 3/86 (3%)
Query 41 GPDAPPNWRELRSHLQREGR---VYKEDCLDIIRRVTEITSNEPNLLRLSDPITVVGDIH 97
GP +L H+++ R + ++D + + EI + E N+ + P+T+ GDIH
Sbjct 22 GPSDQLTTHDLDRHIEKLMRCELIAEQDVKTLCAKAREILAEEGNVQVIDSPVTICGDIH 81
Query 98 GQYYDLLKLLDVGGDPDTTQYLFLGD 123
GQ+YDL++L VGG T YLFLGD
Sbjct 82 GQFYDLMELFKVGGPVPNTNYLFLGD 107
> tgo:TGME49_015170 serine/threonine protein phosphatase, putative
(EC:3.1.3.16); K04382 protein phosphatase 2 (formerly 2A),
catalytic subunit [EC:3.1.3.16]
Length=552
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 28/45 (62%), Positives = 34/45 (75%), Gaps = 0/45 (0%)
Query 79 NEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
+EPN + + P+TV GDIHGQ+YDLL+L VGG P T YLFLGD
Sbjct 250 SEPNCVTVPTPVTVAGDIHGQFYDLLELFRVGGKPPNTSYLFLGD 294
> cel:Y49E10.3 pph-4.2; Protein PHosphatase family member (pph-4.2)
Length=321
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 28/55 (50%), Positives = 37/55 (67%), Gaps = 0/55 (0%)
Query 69 IIRRVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
I +V EI E N+ + P+T+ GDIHGQ++DL++L VGG P T YLFLGD
Sbjct 38 ICAKVREILIEEANVQVIDTPVTICGDIHGQFHDLMELFRVGGSPPNTNYLFLGD 92
> ath:AT4G26720 PPX1; PPX1 (PROTEIN PHOSPHATASE X 1); protein
serine/threonine phosphatase; K01090 protein phosphatase [EC:3.1.3.16]
Length=305
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 26/52 (50%), Positives = 36/52 (69%), Gaps = 0/52 (0%)
Query 72 RVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
+ EI E N+ R+ P+T+ GDIHGQ+YD+++L VGGD T YLF+GD
Sbjct 28 KAMEILVEESNVQRVDAPVTLCGDIHGQFYDMMELFKVGGDCPKTNYLFMGD 79
> ath:AT5G55260 PPX2; PPX2 (PROTEIN PHOSPHATASE X 2); protein
serine/threonine phosphatase; K01090 protein phosphatase [EC:3.1.3.16]
Length=305
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 33/72 (45%), Positives = 42/72 (58%), Gaps = 5/72 (6%)
Query 52 RSHLQREGRVYKEDCLDIIRRVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGG 111
R +E V K CL + EI E N+ R+ P+T+ GDIHGQ+YD+ +L VGG
Sbjct 13 RCEALKESEV-KALCL----KAMEILVEESNVQRVDAPVTICGDIHGQFYDMKELFKVGG 67
Query 112 DPDTTQYLFLGD 123
D T YLFLGD
Sbjct 68 DCPKTNYLFLGD 79
> bbo:BBOV_I000740 16.m00696; serine/threonine protein phosphatase
PP2A catalytic subunit; K04382 protein phosphatase 2 (formerly
2A), catalytic subunit [EC:3.1.3.16]
Length=323
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 30/61 (49%), Positives = 39/61 (63%), Gaps = 0/61 (0%)
Query 63 KEDCLDIIRRVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLG 122
++ L + R EI S E N++ +S PITVVGDIHGQ YDL +L + G T +LFLG
Sbjct 32 EQAVLKLCNRAREILSKETNVISVSAPITVVGDIHGQLYDLKELFRIAGTAPNTNFLFLG 91
Query 123 D 123
D
Sbjct 92 D 92
> pfa:PF14_0142 PP1; serine/threonine protein phosphatase; K06269
protein phosphatase 1, catalytic subunit [EC:3.1.3.16]
Length=304
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 29/49 (59%), Positives = 34/49 (69%), Gaps = 0/49 (0%)
Query 75 EITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
EI N+P LL L PI + GDIHGQ+YDLL+L + GG P YLFLGD
Sbjct 42 EIFLNQPILLELEAPIKICGDIHGQFYDLLRLFEYGGFPPDANYLFLGD 90
> tgo:TGME49_110700 serine/threonine protein phosphatase, putative
(EC:3.1.3.16); K06269 protein phosphatase 1, catalytic
subunit [EC:3.1.3.16]
Length=306
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 29/55 (52%), Positives = 36/55 (65%), Gaps = 0/55 (0%)
Query 69 IIRRVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
+ + EI ++P LL L PI + GDIHGQYYDLL+L + GG P YLFLGD
Sbjct 37 LCHKSREIFISQPILLELEAPIKICGDIHGQYYDLLRLFEYGGFPPEANYLFLGD 91
> dre:571753 ppp1cc, fe06g11, fi22e08, wu:fe06g11, wu:fi22e08;
protein phosphatase 1, catalytic subunit, gamma isoform; K06269
protein phosphatase 1, catalytic subunit [EC:3.1.3.16]
Length=242
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 28/49 (57%), Positives = 35/49 (71%), Gaps = 0/49 (0%)
Query 75 EITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
EI ++P LL L P+ + GDIHGQYYDLL+L + GG P + YLFLGD
Sbjct 74 EIFLSQPILLELEAPLKICGDIHGQYYDLLRLFEYGGYPPESNYLFLGD 122
> ath:AT2G29400 TOPP1; TOPP1 (TYPE ONE PROTEIN PHOSPHATASE 1);
protein serine/threonine phosphatase
Length=318
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 29/49 (59%), Positives = 33/49 (67%), Gaps = 0/49 (0%)
Query 75 EITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
EI +PNLL L PI + GDIHGQY DLL+L + GG P YLFLGD
Sbjct 57 EIFLQQPNLLELEAPIKICGDIHGQYSDLLRLFEYGGFPPEANYLFLGD 105
> xla:432334 ppp4c, MGC78774; protein phosphatase 4, catalytic
subunit (EC:3.1.3.16); K01090 protein phosphatase [EC:3.1.3.16]
Length=307
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 30/70 (42%), Positives = 42/70 (60%), Gaps = 0/70 (0%)
Query 54 HLQREGRVYKEDCLDIIRRVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDP 113
L+R + + + + + EI E N+ R+ P+TV GDIHGQ+YDL +L VGGD
Sbjct 13 QLRRCELIKESEVKALCAKAREILVEESNVQRVDSPVTVCGDIHGQFYDLKELFRVGGDV 72
Query 114 DTTQYLFLGD 123
T YLF+GD
Sbjct 73 PETNYLFMGD 82
> pfa:PFC0595c serine/threonine protein phosphatase, putative;
K01090 protein phosphatase [EC:3.1.3.16]
Length=308
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 27/55 (49%), Positives = 37/55 (67%), Gaps = 0/55 (0%)
Query 69 IIRRVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
+ +RV EI E N+ + P+ + GDIHGQ++DLL+L DVGGD Y+FLGD
Sbjct 29 VCQRVKEILVEENNVQSIKPPVIICGDIHGQFFDLLELFDVGGDIMNNDYIFLGD 83
> mmu:56420 Ppp4c, 1110002D08Rik, AU016079, Ppx; protein phosphatase
4, catalytic subunit (EC:3.1.3.16); K01090 protein phosphatase
[EC:3.1.3.16]
Length=307
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 30/70 (42%), Positives = 42/70 (60%), Gaps = 0/70 (0%)
Query 54 HLQREGRVYKEDCLDIIRRVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDP 113
L+R + + + + + EI E N+ R+ P+TV GDIHGQ+YDL +L VGGD
Sbjct 13 QLRRCELIKESEVKALCAKAREILVEESNVQRVDSPVTVCGDIHGQFYDLKELFRVGGDV 72
Query 114 DTTQYLFLGD 123
T YLF+GD
Sbjct 73 PETNYLFMGD 82
> hsa:5531 PPP4C, PP4, PP4C, PPH3, PPP4, PPX; protein phosphatase
4, catalytic subunit (EC:3.1.3.16); K01090 protein phosphatase
[EC:3.1.3.16]
Length=307
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 30/70 (42%), Positives = 42/70 (60%), Gaps = 0/70 (0%)
Query 54 HLQREGRVYKEDCLDIIRRVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDP 113
L+R + + + + + EI E N+ R+ P+TV GDIHGQ+YDL +L VGGD
Sbjct 13 QLRRCELIKESEVKALCAKAREILVEESNVQRVDSPVTVCGDIHGQFYDLKELFRVGGDV 72
Query 114 DTTQYLFLGD 123
T YLF+GD
Sbjct 73 PETNYLFMGD 82
> dre:562705 ppp4cb, PP4C-B, zgc:172126; protein phosphatase 4
(formerly X), catalytic subunit b (EC:3.1.3.16); K01090 protein
phosphatase [EC:3.1.3.16]
Length=307
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 30/70 (42%), Positives = 42/70 (60%), Gaps = 0/70 (0%)
Query 54 HLQREGRVYKEDCLDIIRRVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDP 113
L+R + + + + + EI E N+ R+ P+TV GDIHGQ+YDL +L VGGD
Sbjct 13 QLRRCELIKENEVKALCAKAREILVEESNVQRVDSPVTVCGDIHGQFYDLKELFRVGGDV 72
Query 114 DTTQYLFLGD 123
T YLF+GD
Sbjct 73 PETNYLFMGD 82
> cel:F56C9.1 gsp-2; yeast Glc Seven-like Phosphatases family
member (gsp-2); K06269 protein phosphatase 1, catalytic subunit
[EC:3.1.3.16]
Length=333
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 27/55 (49%), Positives = 38/55 (69%), Gaps = 0/55 (0%)
Query 69 IIRRVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
+ ++ EI ++P LL L P+ + GD+HGQYYDLL+L + GG P + YLFLGD
Sbjct 37 LCQKSREIFLSQPILLELEAPLKICGDVHGQYYDLLRLFEYGGFPPESNYLFLGD 91
> bbo:BBOV_III006130 17.m07543; Ser/Thr protein phosphatase family
protein; K06269 protein phosphatase 1, catalytic subunit
[EC:3.1.3.16]
Length=301
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 29/49 (59%), Positives = 34/49 (69%), Gaps = 0/49 (0%)
Query 75 EITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
EI +E LL L PI + GDIHGQYYDLL+L + GG P + YLFLGD
Sbjct 40 EIFLSESVLLELEAPIKICGDIHGQYYDLLRLFEYGGFPPASNYLFLGD 88
> xla:380598 ppp1cc, MGC64327, PP-1G-B, ppp1cc-B, xPP1-zeta; protein
phosphatase 1, catalytic subunit, gamma isoform (EC:3.1.3.16);
K06269 protein phosphatase 1, catalytic subunit [EC:3.1.3.16]
Length=323
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 28/49 (57%), Positives = 35/49 (71%), Gaps = 0/49 (0%)
Query 75 EITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
EI ++P LL L P+ + GDIHGQYYDLL+L + GG P + YLFLGD
Sbjct 44 EIFLSQPILLELEAPLKICGDIHGQYYDLLRLFEYGGFPPESNYLFLGD 92
> xla:397767 ppp1cc, MGC85040, ppp1cc-a, ppp1g; protein phosphatase
1, catalytic subunit, gamma isozyme (EC:3.1.3.16); K06269
protein phosphatase 1, catalytic subunit [EC:3.1.3.16]
Length=323
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 28/49 (57%), Positives = 35/49 (71%), Gaps = 0/49 (0%)
Query 75 EITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
EI ++P LL L P+ + GDIHGQYYDLL+L + GG P + YLFLGD
Sbjct 44 EIFLSQPILLELEAPLKICGDIHGQYYDLLRLFEYGGFPPESNYLFLGD 92
> mmu:19047 Ppp1cc, PP1, dis2m1; protein phosphatase 1, catalytic
subunit, gamma isoform (EC:3.1.3.16); K06269 protein phosphatase
1, catalytic subunit [EC:3.1.3.16]
Length=323
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 28/49 (57%), Positives = 35/49 (71%), Gaps = 0/49 (0%)
Query 75 EITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
EI ++P LL L P+ + GDIHGQYYDLL+L + GG P + YLFLGD
Sbjct 44 EIFLSQPILLELEAPLKICGDIHGQYYDLLRLFEYGGFPPESNYLFLGD 92
> hsa:5501 PPP1CC, PP1gamma, PPP1G; protein phosphatase 1, catalytic
subunit, gamma isozyme (EC:3.1.3.16); K06269 protein
phosphatase 1, catalytic subunit [EC:3.1.3.16]
Length=323
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 28/49 (57%), Positives = 35/49 (71%), Gaps = 0/49 (0%)
Query 75 EITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
EI ++P LL L P+ + GDIHGQYYDLL+L + GG P + YLFLGD
Sbjct 44 EIFLSQPILLELEAPLKICGDIHGQYYDLLRLFEYGGFPPESNYLFLGD 92
> cpv:cgd1_2360 protein phosphatase 4 (formerly X), catalytic
subunit; Protein phosphatase 4, catalytic subunit ; K01090 protein
phosphatase [EC:3.1.3.16]
Length=304
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 26/55 (47%), Positives = 37/55 (67%), Gaps = 0/55 (0%)
Query 69 IIRRVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
+ + EI E N+ R+ P+T+ GDIHGQ++DL++L VGG+ T YLFLGD
Sbjct 25 LCMKAREILVEEANVQRIDTPVTICGDIHGQFFDLMELFKVGGELPDTNYLFLGD 79
> cpv:cgd5_4070 hypothetical protein ; K04382 protein phosphatase
2 (formerly 2A), catalytic subunit [EC:3.1.3.16]
Length=239
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 26/55 (47%), Positives = 38/55 (69%), Gaps = 0/55 (0%)
Query 69 IIRRVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
+ ++ EI NE N+ ++ P+T+ GDIHGQ++DLL+L +GG P YLFLGD
Sbjct 62 LCSKLKEILINESNVQKVRCPVTIAGDIHGQFFDLLELFRIGGMPPDVNYLFLGD 116
> hsa:5499 PPP1CA, MGC15877, MGC1674, PP-1A, PP1alpha, PPP1A;
protein phosphatase 1, catalytic subunit, alpha isozyme (EC:3.1.3.16);
K06269 protein phosphatase 1, catalytic subunit [EC:3.1.3.16]
Length=341
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 28/49 (57%), Positives = 35/49 (71%), Gaps = 0/49 (0%)
Query 75 EITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
EI ++P LL L P+ + GDIHGQYYDLL+L + GG P + YLFLGD
Sbjct 55 EIFLSQPILLELEAPLKICGDIHGQYYDLLRLFEYGGFPPESNYLFLGD 103
> mmu:19045 Ppp1ca, Ppp1c, dism2; protein phosphatase 1, catalytic
subunit, alpha isoform (EC:3.1.3.16); K06269 protein phosphatase
1, catalytic subunit [EC:3.1.3.16]
Length=330
Score = 62.0 bits (149), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 28/49 (57%), Positives = 35/49 (71%), Gaps = 0/49 (0%)
Query 75 EITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
EI ++P LL L P+ + GDIHGQYYDLL+L + GG P + YLFLGD
Sbjct 44 EIFLSQPILLELEAPLKICGDIHGQYYDLLRLFEYGGFPPESNYLFLGD 92
> sce:YER133W GLC7, CID1, DIS2; Glc7p (EC:3.1.3.16); K06269 protein
phosphatase 1, catalytic subunit [EC:3.1.3.16]
Length=312
Score = 61.6 bits (148), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 28/55 (50%), Positives = 35/55 (63%), Gaps = 0/55 (0%)
Query 69 IIRRVTEITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
+ + I +P LL L PI + GDIHGQYYDLL+L + GG P + YLFLGD
Sbjct 37 LCSKARSIFIKQPILLELEAPIKICGDIHGQYYDLLRLFEYGGFPPESNYLFLGD 91
> ath:AT3G05580 serine/threonine protein phosphatase, putative;
K01090 protein phosphatase [EC:3.1.3.16]
Length=318
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 28/49 (57%), Positives = 35/49 (71%), Gaps = 0/49 (0%)
Query 75 EITSNEPNLLRLSDPITVVGDIHGQYYDLLKLLDVGGDPDTTQYLFLGD 123
+I ++PNLL L PI + GDIHGQY DLL+L + GG P + YLFLGD
Sbjct 46 QIFLSQPNLLELHAPIRICGDIHGQYQDLLRLFEYGGYPPSANYLFLGD 94
Lambda K H
0.316 0.140 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2003197800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40