bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7390_orf2
Length=84
Score E
Sequences producing significant alignments: (Bits) Value
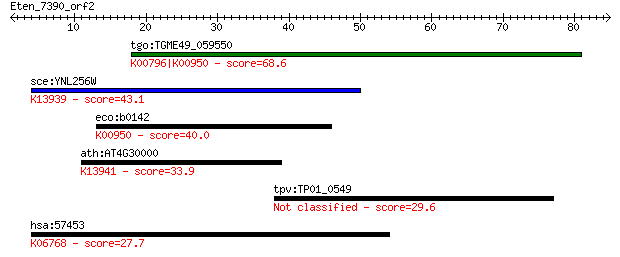
tgo:TGME49_059550 hydroxymethyldihydropterin pyrophosphokinase... 68.6 5e-12
sce:YNL256W FOL1; Fol1p (EC:4.1.2.25 2.7.6.3 2.5.1.15); K13939... 43.1 2e-04
eco:b0142 folK, ECK0141, JW0138; 2-amino-4-hydroxy-6-hydroxyme... 40.0 0.002
ath:AT4G30000 dihydropterin pyrophosphokinase, putative / dihy... 33.9 0.12
tpv:TP01_0549 hypothetical protein 29.6 2.8
hsa:57453 DSCAML1, KIAA1132; Down syndrome cell adhesion molec... 27.7 9.7
> tgo:TGME49_059550 hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate
synthase (EC:2.7.6.3 2.5.1.15); K00796 dihydropteroate
synthase [EC:2.5.1.15]; K00950 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine
diphosphokinase [EC:2.7.6.3]
Length=748
Score = 68.6 bits (166), Expect = 5e-12, Method: Composition-based stats.
Identities = 29/63 (46%), Positives = 43/63 (68%), Gaps = 0/63 (0%)
Query 18 LTLPHPRLAQRNFILFPLCDIKPDLVHPTERETMKNLILKNMRRREEALRHSASSSGETS 77
LTLPHPR+ RNF+LFPLCD+ P+ +HP E ++K L+ N+ RR L ++A + +T
Sbjct 273 LTLPHPRIITRNFVLFPLCDLDPEYIHPVEGMSVKELLRINLERRRIHLLNAAEEARQTQ 332
Query 78 DAA 80
+ A
Sbjct 333 ELA 335
> sce:YNL256W FOL1; Fol1p (EC:4.1.2.25 2.7.6.3 2.5.1.15); K13939
dihydroneopterin aldolase / 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine
diphosphokinase / dihydropteroate synthase
[EC:4.1.2.25 2.7.6.3 2.5.1.15]
Length=824
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/47 (42%), Positives = 30/47 (63%), Gaps = 1/47 (2%)
Query 4 VDSASYSMQVNTTNLTLPHPRLAQRNFILFPLCD-IKPDLVHPTERE 49
++SA + VN +L +PHPR+ +R F+L PLC+ I P +HP E
Sbjct 399 LNSAGEDIIVNEPDLNIPHPRMLERTFVLEPLCELISPVHLHPVTAE 445
> eco:b0142 folK, ECK0141, JW0138; 2-amino-4-hydroxy-6-hydroxymethyldihyropteridine
pyrophosphokinase (EC:2.7.6.3); K00950
2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase
[EC:2.7.6.3]
Length=159
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 16/33 (48%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 13 VNTTNLTLPHPRLAQRNFILFPLCDIKPDLVHP 45
+NT LT+PH + R F+L+PL +I P+LV P
Sbjct 107 INTERLTVPHYDMKNRGFMLWPLFEIAPELVFP 139
> ath:AT4G30000 dihydropterin pyrophosphokinase, putative / dihydropteroate
synthase, putative / DHPS, putative; K13941 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine
diphosphokinase
/ dihydropteroate synthase [EC:2.7.6.3 2.5.1.15]
Length=554
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 12/28 (42%), Positives = 21/28 (75%), Gaps = 0/28 (0%)
Query 11 MQVNTTNLTLPHPRLAQRNFILFPLCDI 38
M++++ L +PH RL +R+F+L PL D+
Sbjct 188 MRISSDKLIIPHERLWERSFVLAPLVDL 215
> tpv:TP01_0549 hypothetical protein
Length=277
Score = 29.6 bits (65), Expect = 2.8, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 26/43 (60%), Gaps = 4/43 (9%)
Query 38 IKPDLVHPTERETMKNLILKNMRRREEA----LRHSASSSGET 76
+KP+L+H + + N +KN++ RE +RH+ ++SG T
Sbjct 235 LKPNLIHDYTIKNIINSKVKNVKERENKSSSIVRHNTNASGNT 277
> hsa:57453 DSCAML1, KIAA1132; Down syndrome cell adhesion molecule
like 1; K06768 Down syndrome cell adhesion molecule like
1
Length=2113
Score = 27.7 bits (60), Expect = 9.7, Method: Composition-based stats.
Identities = 18/54 (33%), Positives = 27/54 (50%), Gaps = 5/54 (9%)
Query 4 VDSASYSMQVN----TTNLTLPHPRLAQRNFILFPLCDIKPDLVHPTERETMKN 53
V A +S VN T ++L HP L Q L + DI+P +P R+ +K+
Sbjct 1744 VTDAEFSQAVNPQSFCTGVSLHHPTLIQSTGPLIDMSDIRPG-TNPVSRKNVKS 1796
Lambda K H
0.317 0.128 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2040069136
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40