bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7406_orf1
Length=171
Score E
Sequences producing significant alignments: (Bits) Value
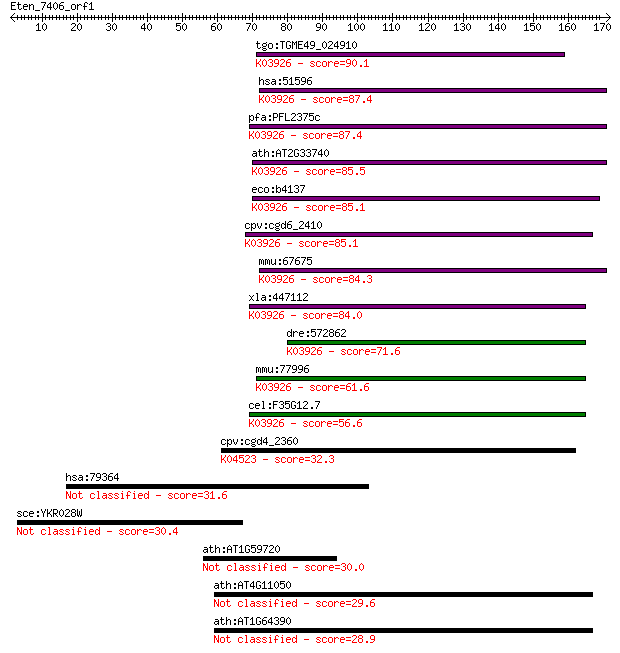
tgo:TGME49_024910 CutA1 divalent ion tolerance domain-containi... 90.1 3e-18
hsa:51596 CUTA, ACHAP, C6orf82, MGC111154; cutA divalent catio... 87.4 2e-17
pfa:PFL2375c CutA, putative; K03926 periplasmic divalent catio... 87.4 2e-17
ath:AT2G33740 CUTA; CUTA; copper ion binding; K03926 periplasm... 85.5 9e-17
eco:b4137 cutA, cutA1, cycY, ECK4131, JW4097; divalent-cation ... 85.1 1e-16
cpv:cgd6_2410 CutA1 divalent ion tolerance protein ; K03926 pe... 85.1 1e-16
mmu:67675 Cuta, 0610039D01Rik, 1810022E02Rik, 1810060C03Rik, 2... 84.3 2e-16
xla:447112 cuta, MGC85327, achap; cutA divalent cation toleran... 84.0 3e-16
dre:572862 MGC174524, MGC63972, cuta; zgc:63972; K03926 peripl... 71.6 1e-12
mmu:77996 D730039F16Rik; RIKEN cDNA D730039F16 gene; K03926 pe... 61.6 1e-09
cel:F35G12.7 hypothetical protein; K03926 periplasmic divalent... 56.6 4e-08
cpv:cgd4_2360 DSK2 like protein with a ubiquitin domain, 2 STI... 32.3 0.81
hsa:79364 ZXDC, DKFZp547N024, FLJ13861, FLJ36477, FLJ37830, MG... 31.6 1.3
sce:YKR028W SAP190; Protein that forms a complex with the Sit4... 30.4 3.3
ath:AT1G59720 CRR28; CRR28 (CHLORORESPIRATORY REDUCTION28); en... 30.0 4.0
ath:AT4G11050 AtGH9C3 (Arabidopsis thaliana glycosyl hydrolase... 29.6 5.1
ath:AT1G64390 AtGH9C2 (Arabidopsis thaliana glycosyl hydrolase... 28.9 8.7
> tgo:TGME49_024910 CutA1 divalent ion tolerance domain-containing
protein ; K03926 periplasmic divalent cation tolerance
protein
Length=148
Score = 90.1 bits (222), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 39/88 (44%), Positives = 60/88 (68%), Gaps = 0/88 (0%)
Query 71 VGFSTASSSEEARSIAENLIVKKLAACVQIVPAVESVYEWKGKLVKSPEVLMIIKAQRQH 130
V + T +A +A L+ +LAACV IVP + S+YEW+GK+ K EVL+I+K +++
Sbjct 18 VAYVTCKDKTQAEEVASKLVENRLAACVNIVPGITSIYEWEGKMEKDEEVLLIVKTRKEL 77
Query 131 AQAVVQAIKEKHSYEVPEVVFTEIVDGN 158
A VV A+++ HSY+VPEV+F ++ GN
Sbjct 78 AAEVVAAVRKWHSYDVPEVIFLDVAGGN 105
> hsa:51596 CUTA, ACHAP, C6orf82, MGC111154; cutA divalent cation
tolerance homolog (E. coli); K03926 periplasmic divalent
cation tolerance protein
Length=198
Score = 87.4 bits (215), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 41/99 (41%), Positives = 60/99 (60%), Gaps = 0/99 (0%)
Query 72 GFSTASSSEEARSIAENLIVKKLAACVQIVPAVESVYEWKGKLVKSPEVLMIIKAQRQHA 131
F T + + A+ IA ++ K+LAACV ++P + S+YEWKGK+ + EVLM+IK Q
Sbjct 90 AFVTCPNEKVAKEIARAVVEKRLAACVNLIPQITSIYEWKGKIEEDSEVLMMIKTQSSLV 149
Query 132 QAVVQAIKEKHSYEVPEVVFTEIVDGNADYLQWARAATQ 170
A+ ++ H YEV EV+ + GN YLQW R T+
Sbjct 150 PALTDFVRSVHPYEVAEVIALPVEQGNFPYLQWVRQVTE 188
> pfa:PFL2375c CutA, putative; K03926 periplasmic divalent cation
tolerance protein
Length=159
Score = 87.4 bits (215), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 38/102 (37%), Positives = 61/102 (59%), Gaps = 0/102 (0%)
Query 69 LVVGFSTASSSEEARSIAENLIVKKLAACVQIVPAVESVYEWKGKLVKSPEVLMIIKAQR 128
+V + T S E A I+ L+ +KL +CV ++P + S+Y WKG++ K EVLM+IK ++
Sbjct 56 FIVVYVTTPSKEVAEKISYVLLEEKLVSCVNVIPGILSLYHWKGEIAKDNEVLMMIKTKK 115
Query 129 QHAQAVVQAIKEKHSYEVPEVVFTEIVDGNADYLQWARAATQ 170
+V+ +K H YE+PEV+ I G+ DYL W + +
Sbjct 116 HLFDEIVKLVKSNHPYEIPEVIAVPIEYGSKDYLDWVNNSVK 157
> ath:AT2G33740 CUTA; CUTA; copper ion binding; K03926 periplasmic
divalent cation tolerance protein
Length=182
Score = 85.5 bits (210), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 38/101 (37%), Positives = 64/101 (63%), Gaps = 0/101 (0%)
Query 70 VVGFSTASSSEEARSIAENLIVKKLAACVQIVPAVESVYEWKGKLVKSPEVLMIIKAQRQ 129
+V + T + E + +A +++ +KLAACV IVP +ESVYEW+GK+ E L+IIK ++
Sbjct 81 IVVYVTVPNREAGKKLANSIVQEKLAACVNIVPGIESVYEWEGKVQSDSEELLIIKTRQS 140
Query 130 HAQAVVQAIKEKHSYEVPEVVFTEIVDGNADYLQWARAATQ 170
+ + + + H Y+VPEV+ I G+ YL+W + +T+
Sbjct 141 LLEPLTEHVNANHEYDVPEVIALPITGGSDKYLEWLKNSTR 181
> eco:b4137 cutA, cutA1, cycY, ECK4131, JW4097; divalent-cation
tolerance protein, copper sensitivity; K03926 periplasmic
divalent cation tolerance protein
Length=112
Score = 85.1 bits (209), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 37/99 (37%), Positives = 58/99 (58%), Gaps = 0/99 (0%)
Query 70 VVGFSTASSSEEARSIAENLIVKKLAACVQIVPAVESVYEWKGKLVKSPEVLMIIKAQRQ 129
VV TA A+ +A ++ +KLAAC ++P S+Y W+GKL + EV MI+K
Sbjct 12 VVVLCTAPDEATAQDLAAKVLAEKLAACATLIPGATSLYYWEGKLEQEYEVQMILKTTVS 71
Query 130 HAQAVVQAIKEKHSYEVPEVVFTEIVDGNADYLQWARAA 168
H QA+++ +K H Y+ PE++ + G+ DYL W A+
Sbjct 72 HQQALLECLKSHHPYQTPELLVLPVTHGDTDYLSWLNAS 110
> cpv:cgd6_2410 CutA1 divalent ion tolerance protein ; K03926
periplasmic divalent cation tolerance protein
Length=116
Score = 85.1 bits (209), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 35/99 (35%), Positives = 67/99 (67%), Gaps = 0/99 (0%)
Query 68 GLVVGFSTASSSEEARSIAENLIVKKLAACVQIVPAVESVYEWKGKLVKSPEVLMIIKAQ 127
+++ + +A + +EA SIA+ L+ ++L ACV I+P+V S+Y++KG++ EV++++K
Sbjct 13 NIILIYISAPNQDEATSIAKTLVDEELCACVSIIPSVRSIYKFKGQVHDENEVMLLVKTT 72
Query 128 RQHAQAVVQAIKEKHSYEVPEVVFTEIVDGNADYLQWAR 166
Q + + + E HSYE+PE++ T++V GN +Y+ W
Sbjct 73 SQLFTTLKEKVTEIHSYELPEIIATKVVYGNENYINWVN 111
> mmu:67675 Cuta, 0610039D01Rik, 1810022E02Rik, 1810060C03Rik,
2700094G22Rik, AI326454; cutA divalent cation tolerance homolog
(E. coli); K03926 periplasmic divalent cation tolerance
protein
Length=177
Score = 84.3 bits (207), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 39/99 (39%), Positives = 59/99 (59%), Gaps = 0/99 (0%)
Query 72 GFSTASSSEEARSIAENLIVKKLAACVQIVPAVESVYEWKGKLVKSPEVLMIIKAQRQHA 131
F T + + A+ IA ++ K+LAACV ++P + S+YEWKGK+ + EVLM+IK Q
Sbjct 69 AFVTCPNEKVAKEIARAVVEKRLAACVNLIPQITSIYEWKGKIEEDSEVLMMIKTQSSLV 128
Query 132 QAVVQAIKEKHSYEVPEVVFTEIVDGNADYLQWARAATQ 170
A+ + ++ H YEV EV+ + GN YL W T+
Sbjct 129 PALTEFVRSVHPYEVAEVIALPVEQGNPPYLHWVHQVTE 167
> xla:447112 cuta, MGC85327, achap; cutA divalent cation tolerance
homolog; K03926 periplasmic divalent cation tolerance protein
Length=113
Score = 84.0 bits (206), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 39/96 (40%), Positives = 58/96 (60%), Gaps = 0/96 (0%)
Query 69 LVVGFSTASSSEEARSIAENLIVKKLAACVQIVPAVESVYEWKGKLVKSPEVLMIIKAQR 128
L + T + A+ IA L+ +KLAACV ++P + S+YEWKGKL + EVL++IK +
Sbjct 11 LSAAYVTCPNDTVAKDIARGLVERKLAACVNVIPQITSIYEWKGKLEEDTEVLLMIKTRS 70
Query 129 QHAQAVVQAIKEKHSYEVPEVVFTEIVDGNADYLQW 164
A+ + ++ H YEV EV+ I GN YL+W
Sbjct 71 SKVSALTEYVRSVHPYEVCEVISLPIEQGNPPYLKW 106
> dre:572862 MGC174524, MGC63972, cuta; zgc:63972; K03926 periplasmic
divalent cation tolerance protein
Length=150
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 29/85 (34%), Positives = 52/85 (61%), Gaps = 0/85 (0%)
Query 80 EEARSIAENLIVKKLAACVQIVPAVESVYEWKGKLVKSPEVLMIIKAQRQHAQAVVQAIK 139
+ AR I ++ K+LAACV I P ++Y WKG++ + E+L++++ + Q ++ I
Sbjct 58 QTARDIGRIIMEKRLAACVNIFPRTATMYYWKGEIRDATEILLLVRTKTSLVQRLMTYIT 117
Query 140 EKHSYEVPEVVFTEIVDGNADYLQW 164
H Y++PE++ I DG+ YL+W
Sbjct 118 AIHPYDIPEIITFPINDGSQHYLKW 142
> mmu:77996 D730039F16Rik; RIKEN cDNA D730039F16 gene; K03926
periplasmic divalent cation tolerance protein
Length=156
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 53/94 (56%), Gaps = 0/94 (0%)
Query 71 VGFSTASSSEEARSIAENLIVKKLAACVQIVPAVESVYEWKGKLVKSPEVLMIIKAQRQH 130
+ F + + AR IA ++ KK+A+ V I+P S+Y WKG++ + EV ++IK +
Sbjct 55 IVFVNCPNEQIARDIARAILDKKMASSVNILPKTSSLYFWKGEIEEGIEVSLLIKTKTSK 114
Query 131 AQAVVQAIKEKHSYEVPEVVFTEIVDGNADYLQW 164
+ ++ H +E+PEV + G+A +L+W
Sbjct 115 VSRLFAYMRLAHPFEIPEVFSIPMDQGDARFLRW 148
> cel:F35G12.7 hypothetical protein; K03926 periplasmic divalent
cation tolerance protein
Length=115
Score = 56.6 bits (135), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 31/97 (31%), Positives = 52/97 (53%), Gaps = 2/97 (2%)
Query 69 LVVGFSTASSSEEARSIAENLIVKKLAACVQIVPAVESVYEWKGKLVKSPEVLMIIKAQR 128
+VV + TA S E A ++A + + LAAC ++P V SVY+W+GK+ + E ++I+K
Sbjct 8 MVVAYVTAPSKEVAMTVARTTVTEALAACANVIPEVTSVYKWQGKIEEDQEHVVILKTVE 67
Query 129 QHAQAVVQAIKEKHSYEVPEVVFTEIVDG-NADYLQW 164
+ + ++ H E P FT +D D+ W
Sbjct 68 SKVEELSARVRSLHPAETP-CFFTLAIDKITPDFGGW 103
> cpv:cgd4_2360 DSK2 like protein with a ubiquitin domain, 2 STI1
motifs and a UBA domain at its C-terminus ; K04523 ubiquilin
Length=402
Score = 32.3 bits (72), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 26/104 (25%), Positives = 48/104 (46%), Gaps = 8/104 (7%)
Query 61 IAMASTEGLVVGFSTASSSEEARSIAENLIVKKLAACV---QIVPAVESVYEWKGKLVKS 117
+ MAS + + + F + ++ S+ N IVK L + +P+ + +KG+++K
Sbjct 46 LKMASIDEISIVFKVSGGTQFNISVPRNTIVKDLKDKISEPSNIPSSQQRLIYKGRILKD 105
Query 118 PEVLMIIKAQRQHAQAVVQAIKEKHSYEVPEVVFTEIVDGNADY 161
+ L +K + H +V K +V T I+D ADY
Sbjct 106 SDSLDGMKVESGHTMHLV-----KSGVQVENQKPTNILDQAADY 144
> hsa:79364 ZXDC, DKFZp547N024, FLJ13861, FLJ36477, FLJ37830,
MGC11349, ZXDL; ZXD family zinc finger C
Length=710
Score = 31.6 bits (70), Expect = 1.3, Method: Composition-based stats.
Identities = 22/86 (25%), Positives = 39/86 (45%), Gaps = 3/86 (3%)
Query 17 PLIKGQGSSPRENGLVASNSFNSSTYTDRECSRALQMENQGTQRIAMASTEGLVVGFSTA 76
PL+ G ++ + G + + + + C AL M+N +A+ S L +T
Sbjct 581 PLVLGTAATVLQQGSFSVDDVQTVSAGALGCLVALPMKNLSDDPLALTSNSNLAAHITTP 640
Query 77 SSSEEARSIAENLIVKKLAACVQIVP 102
+SS R EN V +L A +++ P
Sbjct 641 TSSSTPR---ENASVPELLAPIKVEP 663
> sce:YKR028W SAP190; Protein that forms a complex with the Sit4p
protein phosphatase and is required for its function; member
of a family of similar proteins including Sap4p, Sap155p,
and Sap185p
Length=1098
Score = 30.4 bits (67), Expect = 3.3, Method: Composition-based stats.
Identities = 22/72 (30%), Positives = 36/72 (50%), Gaps = 18/72 (25%)
Query 3 RERERERERERETPPLIKGQGSSPRENGLVASNSFNSSTYTDRECSR--------ALQME 54
+ER+ ER +E ET S+ +EN + + N S+Y D++C ALQ+
Sbjct 509 QERDIERAKELET--------STEKEN--ITAIVDNKSSYYDKDCVEKDITENLGALQIN 558
Query 55 NQGTQRIAMAST 66
NQG++ + T
Sbjct 559 NQGSEEDELNDT 570
> ath:AT1G59720 CRR28; CRR28 (CHLORORESPIRATORY REDUCTION28);
endonuclease
Length=638
Score = 30.0 bits (66), Expect = 4.0, Method: Composition-based stats.
Identities = 12/38 (31%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 56 QGTQRIAMASTEGLVVGFSTASSSEEARSIAENLIVKK 93
QG Q+ +AS +++GF+T +EEA + + ++ K+
Sbjct 313 QGMQKRDLASWNAMILGFATHGRAEEAMNFFDRMVDKR 350
> ath:AT4G11050 AtGH9C3 (Arabidopsis thaliana glycosyl hydrolase
9C3); carbohydrate binding / catalytic/ hydrolase, hydrolyzing
O-glycosyl compounds (EC:3.2.1.4)
Length=626
Score = 29.6 bits (65), Expect = 5.1, Method: Composition-based stats.
Identities = 26/108 (24%), Positives = 45/108 (41%), Gaps = 6/108 (5%)
Query 59 QRIAMASTEGLVVGFSTASSSEEARSIAENLIVKKLAACVQIVPAVESVYEWKGKLVKSP 118
QR++ S GL G S+ A + + L + S+ E+ G+L +
Sbjct 50 QRVSWRSHSGLYDGKSSGVDLVGGYYDAGDNVKFGLPMAFTVTTMCWSIIEYGGQLESNG 109
Query 119 EVLMIIKAQRQHAQAVVQAIKEKHSYEVPEVVFTEIVDGNADYLQWAR 166
E+ I A + ++A E P V++ E+ DG +D+ W R
Sbjct 110 ELGHAIDAVKWGTDYFIKAHPE------PNVLYGEVGDGKSDHYCWQR 151
> ath:AT1G64390 AtGH9C2 (Arabidopsis thaliana glycosyl hydrolase
9C2); carbohydrate binding / catalytic/ hydrolase, hydrolyzing
O-glycosyl compounds (EC:3.2.1.4)
Length=620
Score = 28.9 bits (63), Expect = 8.7, Method: Composition-based stats.
Identities = 27/108 (25%), Positives = 44/108 (40%), Gaps = 6/108 (5%)
Query 59 QRIAMASTEGLVVGFSTASSSEEARSIAENLIVKKLAACVQIVPAVESVYEWKGKLVKSP 118
QR+ S GL G S+ + A + + L + SV E+ +L +
Sbjct 49 QRVTWRSHSGLTDGKSSGVNLVGGYYDAGDNVKFGLPMAFTVTMMAWSVIEYGNQLQANG 108
Query 119 EVLMIIKAQRQHAQAVVQAIKEKHSYEVPEVVFTEIVDGNADYLQWAR 166
E+ I A + ++A E P V++ E+ DGN D+ W R
Sbjct 109 ELGNSIDAIKWGTDYFIKAHPE------PNVLYGEVGDGNTDHYCWQR 150
Lambda K H
0.311 0.125 0.340
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4276754328
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40