bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7413_orf1
Length=123
Score E
Sequences producing significant alignments: (Bits) Value
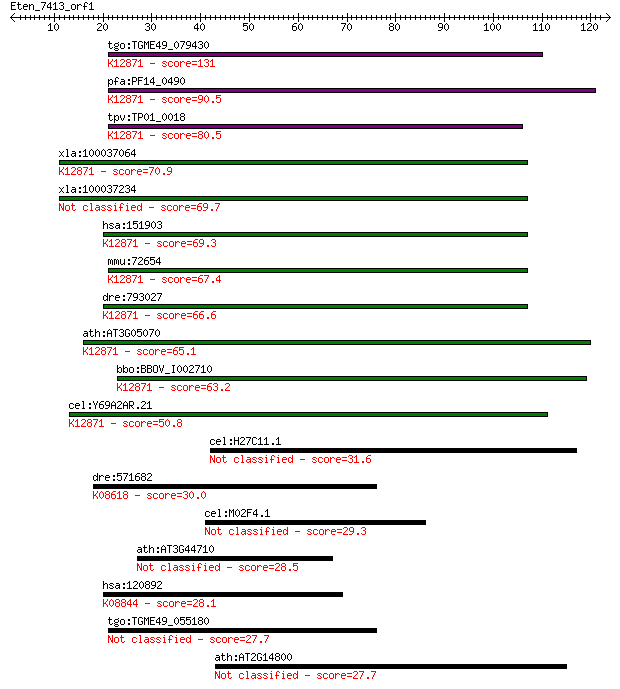
tgo:TGME49_079430 hypothetical protein ; K12871 coiled-coil do... 131 6e-31
pfa:PF14_0490 conserved Plasmodium protein, unknown function; ... 90.5 1e-18
tpv:TP01_0018 hypothetical protein; K12871 coiled-coil domain-... 80.5 1e-15
xla:100037064 hypothetical protein LOC100037064; K12871 coiled... 70.9 1e-12
xla:100037234 ccdc12; coiled-coil domain containing 12 69.7
hsa:151903 CCDC12, FLJ39430, FLJ40801, MGC23918; coiled-coil d... 69.3 3e-12
mmu:72654 Ccdc12, 2700094L05Rik, C76605; coiled-coil domain co... 67.4 1e-11
dre:793027 ccdc12, MGC101038, zgc:101038; coiled-coil domain c... 66.6 2e-11
ath:AT3G05070 hypothetical protein; K12871 coiled-coil domain-... 65.1 5e-11
bbo:BBOV_I002710 19.m02067; hypothetical protein; K12871 coile... 63.2 2e-10
cel:Y69A2AR.21 hypothetical protein; K12871 coiled-coil domain... 50.8 9e-07
cel:H27C11.1 nhr-97; Nuclear Hormone Receptor family member (n... 31.6 0.68
dre:571682 adamts3; ADAM metallopeptidase with thrombospondin ... 30.0 1.9
cel:M02F4.1 hypothetical protein 29.3 3.1
ath:AT3G44710 hypothetical protein 28.5 5.4
hsa:120892 LRRK2, AURA17, DARDARIN, PARK8, RIPK7, ROCO2; leuci... 28.1 6.4
tgo:TGME49_055180 ubiquitin carboxyl-terminal hydrolase, putat... 27.7 8.6
ath:AT2G14800 hypothetical protein 27.7 9.0
> tgo:TGME49_079430 hypothetical protein ; K12871 coiled-coil
domain-containing protein 12
Length=152
Score = 131 bits (329), Expect = 6e-31, Method: Compositional matrix adjust.
Identities = 58/89 (65%), Positives = 76/89 (85%), Gaps = 0/89 (0%)
Query 21 IRFRNYVPKDPKLRQFCLPRPSVEELEKQIAKEAQEAVQAAKEEDILSQVVPRRPNWDLK 80
+RFRNYVP+D LR+FCLPRPSVEELEKQI +EA +A+ ++ ED+++Q+ PRRPNWDLK
Sbjct 19 LRFRNYVPRDVVLRRFCLPRPSVEELEKQIDREATDAINSSANEDVVAQIAPRRPNWDLK 78
Query 81 RDVERKITILSRRTDKAIVQLIREKIEQN 109
RDVE+K+ +LSR+TD AIV LIR KI+ +
Sbjct 79 RDVEKKLAVLSRKTDMAIVDLIRMKIKNS 107
> pfa:PF14_0490 conserved Plasmodium protein, unknown function;
K12871 coiled-coil domain-containing protein 12
Length=147
Score = 90.5 bits (223), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 40/100 (40%), Positives = 69/100 (69%), Gaps = 0/100 (0%)
Query 21 IRFRNYVPKDPKLRQFCLPRPSVEELEKQIAKEAQEAVQAAKEEDILSQVVPRRPNWDLK 80
+RF NY+P + +L++ C+P P E+ E ++ KE E + + +IL Q+ + N DLK
Sbjct 9 LRFYNYIPVNKELKKSCIPCPDTEDYEAKLNKEFDEELTKVFQGNILDQINAKDINADLK 68
Query 81 RDVERKITILSRRTDKAIVQLIREKIEQNKKAKELASTEE 120
RD+++K+ ILS++TDKA+VQLI++KI +NK + + +T +
Sbjct 69 RDLKKKLDILSKKTDKAVVQLIKQKINENKNKENITNTND 108
> tpv:TP01_0018 hypothetical protein; K12871 coiled-coil domain-containing
protein 12
Length=146
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 41/88 (46%), Positives = 60/88 (68%), Gaps = 5/88 (5%)
Query 21 IRFRNYVPKDPKLRQFCLPRPSVEE---LEKQIAKEAQEAVQAAKEEDILSQVVPRRPNW 77
I FRNY+P+D LR+ C + S+E+ +E I ++ + + + EDILS V PRR NW
Sbjct 27 IVFRNYIPRDENLRKLC--KSSLEDYSAIENTIDQQIDQTILNYQSEDILSLVRPRRQNW 84
Query 78 DLKRDVERKITILSRRTDKAIVQLIREK 105
DLKR++ RK +LS RTD AI++L+RE+
Sbjct 85 DLKRELNRKRQVLSSRTDAAILRLLRER 112
> xla:100037064 hypothetical protein LOC100037064; K12871 coiled-coil
domain-containing protein 12
Length=157
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 43/104 (41%), Positives = 64/104 (61%), Gaps = 15/104 (14%)
Query 11 AGRPREASPEIRFRNYVPKDPKLRQFCLPRP---SVEELEKQIAKEAQEAVQAAKEEDI- 66
AG E E++ RNY P+D L++ +P+ SVEE + QE ++AAK E I
Sbjct 39 AGEEEEQHRELKLRNYTPQDEVLKERQVPQAKPISVEE-------KVQEQLEAAKPEPII 91
Query 67 ----LSQVVPRRPNWDLKRDVERKITILSRRTDKAIVQLIREKI 106
L+ + PR+P+WDLKRDV +K+ L +RT +AI +LIRE++
Sbjct 92 EEVDLANLAPRKPDWDLKRDVAKKLEKLEKRTQRAIAELIRERL 135
> xla:100037234 ccdc12; coiled-coil domain containing 12
Length=157
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 42/104 (40%), Positives = 65/104 (62%), Gaps = 15/104 (14%)
Query 11 AGRPREASPEIRFRNYVPKDPKLRQFCLPRP---SVEELEKQIAKEAQEAVQAAKEEDI- 66
AG +E E++ RNY P+D L++ +P+ SVEE + +E ++AAK E I
Sbjct 39 AGEEQEQHRELKLRNYTPQDDVLKERQVPQAKPISVEE-------KVKEQLEAAKPEPII 91
Query 67 ----LSQVVPRRPNWDLKRDVERKITILSRRTDKAIVQLIREKI 106
L+ + PR+P+WDLKRDV +K+ L +RT +AI +LIRE++
Sbjct 92 EEVDLANLAPRKPDWDLKRDVAKKLEKLEKRTQRAIAELIRERL 135
> hsa:151903 CCDC12, FLJ39430, FLJ40801, MGC23918; coiled-coil
domain containing 12; K12871 coiled-coil domain-containing
protein 12
Length=179
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 37/92 (40%), Positives = 62/92 (67%), Gaps = 9/92 (9%)
Query 20 EIRFRNYVPKDPKLRQFCLPRPSVEELEKQIAKEAQEAVQAAKEEDI-----LSQVVPRR 74
E+R RNYVP+D L++ +P+ +E+++ +E ++AAK E + L+ + PR+
Sbjct 69 ELRLRNYVPEDEDLKKRRVPQAKPVAVEEKV----KEQLEAAKPEPVIEEVDLANLAPRK 124
Query 75 PNWDLKRDVERKITILSRRTDKAIVQLIREKI 106
P+WDLKRDV +K+ L +RT +AI +LIRE++
Sbjct 125 PDWDLKRDVAKKLEKLKKRTQRAIAELIRERL 156
> mmu:72654 Ccdc12, 2700094L05Rik, C76605; coiled-coil domain
containing 12; K12871 coiled-coil domain-containing protein
12
Length=166
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 36/91 (39%), Positives = 61/91 (67%), Gaps = 9/91 (9%)
Query 21 IRFRNYVPKDPKLRQFCLPRPSVEELEKQIAKEAQEAVQAAKEEDI-----LSQVVPRRP 75
+R RNYVP+D L++ +P+ +E+++ +E ++AAK E + L+ + PR+P
Sbjct 57 LRLRNYVPEDEDLKRRRVPQAKPVAVEEKV----KEQLEAAKPEPVIEEVDLANLAPRKP 112
Query 76 NWDLKRDVERKITILSRRTDKAIVQLIREKI 106
+WDLKRDV +K+ L +RT +AI +LIRE++
Sbjct 113 DWDLKRDVAKKLEKLEKRTQRAIAELIRERL 143
> dre:793027 ccdc12, MGC101038, zgc:101038; coiled-coil domain
containing 12; K12871 coiled-coil domain-containing protein
12
Length=163
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 34/88 (38%), Positives = 58/88 (65%), Gaps = 1/88 (1%)
Query 20 EIRFRNYVPKDPKLRQFCLPRPSVEELEKQIAKEAQEAV-QAAKEEDILSQVVPRRPNWD 78
E++ RNY P+D +L++ +P+ ++ ++ + + A + EE L+ + PR+P+WD
Sbjct 54 ELKLRNYTPEDVELKERQVPKAKPASVDDKVKDQLEAANPEPVIEEVDLANLAPRKPDWD 113
Query 79 LKRDVERKITILSRRTDKAIVQLIREKI 106
LKRDV +K+ L RRT KAI +LIRE++
Sbjct 114 LKRDVAKKLEKLERRTQKAIAELIRERL 141
> ath:AT3G05070 hypothetical protein; K12871 coiled-coil domain-containing
protein 12
Length=144
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 39/106 (36%), Positives = 63/106 (59%), Gaps = 11/106 (10%)
Query 16 EASPEIRFRNYVPKDPKLRQFCLPRPSVEELEKQIAKEAQEAVQAA--KEEDILSQVVPR 73
E P ++FRNYVP+ +L+ L P + + E I A+ A K+ED + P+
Sbjct 42 EDGPAMKFRNYVPQAKELQDGKLAPPELPKFEDPIV-----ALPPAVEKKEDPFVNIAPK 96
Query 74 RPNWDLKRDVERKITILSRRTDKAIVQLIREKIEQNKKAKELASTE 119
+PNWDL+RDV++K+ L RRT KA+ +L +E+ +K +E+A +
Sbjct 97 KPNWDLRRDVQKKLDKLERRTQKAMHKL----MEEQEKEREMAEAD 138
> bbo:BBOV_I002710 19.m02067; hypothetical protein; K12871 coiled-coil
domain-containing protein 12
Length=121
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 40/101 (39%), Positives = 60/101 (59%), Gaps = 13/101 (12%)
Query 23 FRNYVPKDPKLRQFCLPRPSVEELEKQIAKEAQEAVQAAKEE----DILSQVVPRRPNWD 78
F +YVPKD L++F R +E+ +QI ++ A+ EE D+LS V+P + NWD
Sbjct 10 FHHYVPKDENLKRFV--RSDLEDY-RQIEEDIDSAINKIIEEHSNKDVLSLVLPSKQNWD 66
Query 79 LKRDVERKITILSRRTDKAIVQLIREKIEQNK-KAKELAST 118
LKRD+ +L+ RTD AI++L+ QN+ K E+ ST
Sbjct 67 LKRDLRDSRQLLATRTDMAIMKLL-----QNQPKTNEIDST 102
> cel:Y69A2AR.21 hypothetical protein; K12871 coiled-coil domain-containing
protein 12
Length=169
Score = 50.8 bits (120), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 33/104 (31%), Positives = 55/104 (52%), Gaps = 15/104 (14%)
Query 13 RPREASPEIRFRNYVPKDPKLRQFC------LPRPSVEELEKQIAKEAQEAVQAAKEEDI 66
+ RE E FRN+ P D Q + R E L+ + ++A ++V
Sbjct 60 KSREVGRE--FRNHKPDDAVGTQNVDMDLDIVQREITEHLKDVLHEKAIDSVD------- 110
Query 67 LSQVVPRRPNWDLKRDVERKITILSRRTDKAIVQLIREKIEQNK 110
L+ + P++ +WDLKRD+E K+ L RRT KA+ +IR+++ + K
Sbjct 111 LAMLAPKKIDWDLKRDIESKLQKLERRTQKAVATIIRQRLAEGK 154
> cel:H27C11.1 nhr-97; Nuclear Hormone Receptor family member
(nhr-97)
Length=439
Score = 31.6 bits (70), Expect = 0.68, Method: Composition-based stats.
Identities = 22/76 (28%), Positives = 35/76 (46%), Gaps = 1/76 (1%)
Query 42 SVEELEKQIAKEAQEAVQAAKEEDILSQVVPRRPNWD-LKRDVERKITILSRRTDKAIVQ 100
+++ L K +E + AKE +L ++ P+ + L D E I +L R A+ Q
Sbjct 291 TIQSLNKWAQRELKPLCLRAKEIVLLKALIALNPDANGLSSDAESSIRMLRERVHTALFQ 350
Query 101 LIREKIEQNKKAKELA 116
L+ E E A LA
Sbjct 351 LLMENSEPITAASRLA 366
> dre:571682 adamts3; ADAM metallopeptidase with thrombospondin
type 1 motif, 3; K08618 a disintegrin and metalloproteinase
with thrombospondin motifs 2 [EC:3.4.24.14]
Length=1270
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 18/58 (31%), Positives = 28/58 (48%), Gaps = 1/58 (1%)
Query 18 SPEIRFRNYVPKDPKLRQFCLPRPSVEELEKQIAKEAQEAVQAAKEEDILSQVVPRRP 75
S ++ +R P + L F +PR V EK K QE + + +D L Q + R+P
Sbjct 1212 SIDVSYRIVSPNEVALNHF-VPRKRVSFREKTQNKRIQELLAEKRRQDFLLQRLKRKP 1268
> cel:M02F4.1 hypothetical protein
Length=399
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 25/48 (52%), Gaps = 3/48 (6%)
Query 41 PSVEELEKQIAKEAQEAVQAAKEEDILSQVV---PRRPNWDLKRDVER 85
PS + Q+ + E V + E+D+LS+ + P+ P RD+ER
Sbjct 94 PSTHHIGSQVRGSSMEEVLTSDEKDLLSKAINNLPKSPKSKSDRDLER 141
> ath:AT3G44710 hypothetical protein
Length=504
Score = 28.5 bits (62), Expect = 5.4, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 25/50 (50%), Gaps = 10/50 (20%)
Query 27 VPKDPK----------LRQFCLPRPSVEELEKQIAKEAQEAVQAAKEEDI 66
VPK+ K +R+ C+PR S +EL+ + K QE + EE I
Sbjct 251 VPKEEKKSNHLLDCMMIRKICMPRESKQELDVMLKKGKQEVDISMLEEGI 300
> hsa:120892 LRRK2, AURA17, DARDARIN, PARK8, RIPK7, ROCO2; leucine-rich
repeat kinase 2 (EC:2.7.11.1); K08844 leucine-rich
repeat kinase 2 [EC:2.7.11.1]
Length=2527
Score = 28.1 bits (61), Expect = 6.4, Method: Composition-based stats.
Identities = 20/50 (40%), Positives = 27/50 (54%), Gaps = 2/50 (4%)
Query 20 EIRFRNYVPKDPKLRQFCLPRPSVEELEKQIAKE-AQEAVQAAKEEDILS 68
E+ + +P DP C P P VE+L KQ KE QE +A+ DIL+
Sbjct 2085 ELEIQGKLP-DPVKEYGCAPWPMVEKLIKQCLKENPQERPTSAQVFDILN 2133
> tgo:TGME49_055180 ubiquitin carboxyl-terminal hydrolase, putative
(EC:3.1.2.15)
Length=4187
Score = 27.7 bits (60), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 28/55 (50%), Gaps = 7/55 (12%)
Query 21 IRFRNYVPKDPKLRQFCLPRPSVEELEKQIAKEAQEAVQAAKEEDILSQVVPRRP 75
+ FR+++ P LR+ +EE+E+Q EA A + SQV+ R+P
Sbjct 789 VHFRDFLLSLPTLRE-------LEEMERQRPAEASRLSLAPSTPETHSQVLARQP 836
> ath:AT2G14800 hypothetical protein
Length=620
Score = 27.7 bits (60), Expect = 9.0, Method: Composition-based stats.
Identities = 18/72 (25%), Positives = 34/72 (47%), Gaps = 6/72 (8%)
Query 43 VEELEKQIAKEAQEAVQAAKEEDILSQVVPRRPNWDLKRDVERKITILSRRTDKAIVQLI 102
VEE+ KQ+ + + + E D+L + +W+ + R +LSRR + +
Sbjct 306 VEEINKQVVAYGEAVMISLNETDVL------QSSWEHQMFSHRPTRLLSRRKQRKCFKSW 359
Query 103 REKIEQNKKAKE 114
R K +Q K+ +
Sbjct 360 RFKFKQVKEGNK 371
Lambda K H
0.310 0.128 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2003197800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40