bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7425_orf2
Length=172
Score E
Sequences producing significant alignments: (Bits) Value
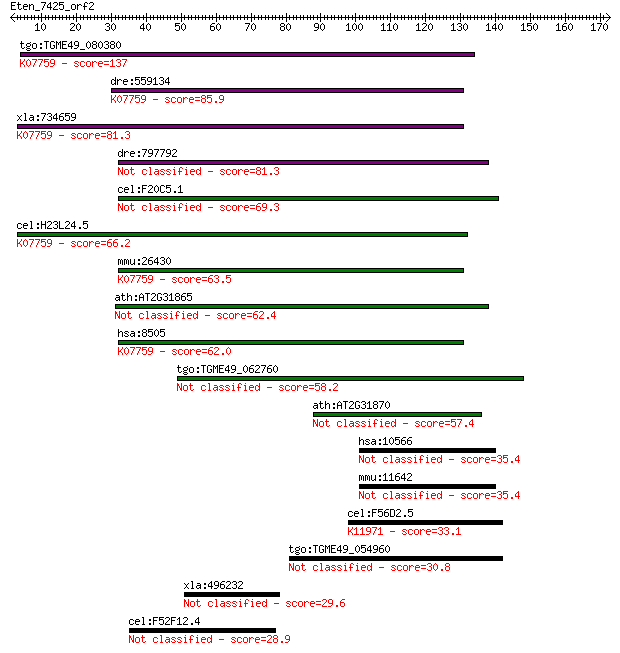
tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143); K0... 137 1e-32
dre:559134 poly(ADP-ribose) glycohydrolase 63 kDa-like; K07759... 85.9 6e-17
xla:734659 parg, MGC115697; poly (ADP-ribose) glycohydrolase; ... 81.3 2e-15
dre:797792 si:dkey-259k14.2 81.3 2e-15
cel:F20C5.1 pme-3; Poly(ADP-ribose) Metabolism Enzyme family m... 69.3 6e-12
cel:H23L24.5 pme-4; Poly(ADP-ribose) Metabolism Enzyme family ... 66.2 5e-11
mmu:26430 Parg, AI413217; poly (ADP-ribose) glycohydrolase (EC... 63.5 4e-10
ath:AT2G31865 poly (ADP-ribose) glycohydrolase (PARG) family p... 62.4 6e-10
hsa:8505 PARG, FLJ54459, FLJ60257, FLJ60456, PARG99; poly (ADP... 62.0 9e-10
tgo:TGME49_062760 poly(ADP-ribose) glycohydrolase, putative 58.2 1e-08
ath:AT2G31870 TEJ; TEJ (Sanskrit for 'bright'); poly(ADP-ribos... 57.4 3e-08
hsa:10566 AKAP3, AKAP110, CT82, FSP95, PRKA3, SOB1; A kinase (... 35.4 0.100
mmu:11642 Akap3, Prka3; A kinase (PRKA) anchor protein 3 35.4
cel:F56D2.5 hypothetical protein; K11971 E3 ubiquitin-protein ... 33.1 0.41
tgo:TGME49_054960 hypothetical protein 30.8 2.6
xla:496232 ppp4r2-a; serine/threonine-protein phosphatase 4 re... 29.6 4.6
cel:F52F12.4 lsl-1; LSY-2-Like family member (lsl-1) 28.9 8.5
> tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143);
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=553
Score = 137 bits (346), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 61/135 (45%), Positives = 90/135 (66%), Gaps = 5/135 (3%)
Query 4 RGYERTFELRAA-----EVPEGCHWSLHGLCPLDGLGRRAATVAAIDALIVRNPNSQYQE 58
+GYE TF++ AA E+P H+S+ G+ PLD LGRR + IDA+ PN QY
Sbjct 375 QGYEWTFQITAAAGDWTELPSNKHFSVQGVVPLDSLGRRNVAIVGIDAVQFHEPNKQYSP 434
Query 59 NKMKREVLKALLGFQGDPFEALLGEPKAPVATGKWGCVIFGGDNQLKTVLQWIAASAAAR 118
+ RE++KA +GF+GDP+E ++ + P+ATG WGC +F GD QLKT++QW+AAS A R
Sbjct 435 VMVNRELMKAAVGFKGDPYELIVSGSRQPIATGLWGCGVFNGDAQLKTLIQWLAASYAGR 494
Query 119 PLQYMAFGDKSLSGM 133
+++ F +KS+ G+
Sbjct 495 SMKFYTFSNKSVDGL 509
> dre:559134 poly(ADP-ribose) glycohydrolase 63 kDa-like; K07759
poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=777
Score = 85.9 bits (211), Expect = 6e-17, Method: Composition-based stats.
Identities = 46/101 (45%), Positives = 58/101 (57%), Gaps = 3/101 (2%)
Query 30 PLDGLGRRAATVAAIDALIVRNPNSQYQENKMKREVLKALLGFQGDPFEALLGEPKAPVA 89
P DG RR + A+DAL RN Q+Q KM RE+ KA GF L + VA
Sbjct 604 PRDGWQRRCTEIVAMDALHYRNFMDQFQPEKMTRELNKAYCGFMRPGVNPL---NLSAVA 660
Query 90 TGKWGCVIFGGDNQLKTVLQWIAASAAARPLQYMAFGDKSL 130
TG WGC FGGD +LK +LQ +AA+ A R + Y FGD++L
Sbjct 661 TGNWGCGAFGGDTRLKALLQLMAAAEAGRDVAYFTFGDEAL 701
> xla:734659 parg, MGC115697; poly (ADP-ribose) glycohydrolase;
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=759
Score = 81.3 bits (199), Expect = 2e-15, Method: Composition-based stats.
Identities = 47/128 (36%), Positives = 61/128 (47%), Gaps = 12/128 (9%)
Query 3 YRGYERTFELRAAEVPEGCHWSLHGLCPLDGLGRRAATVAAIDALIVRNPNSQYQENKMK 62
Y GY T++ E P D RR + AIDA R P Q+ K+K
Sbjct 589 YTGYSETYKWACVHEDES---------PRDEWQRRTTEIVAIDAFHFRRPIDQFVPEKIK 639
Query 63 REVLKALLGFQGDPFEALLGEPKAPVATGKWGCVIFGGDNQLKTVLQWIAASAAARPLQY 122
RE+ KA GF + + VATG WGC FGGD +LK ++Q +AA+ R L Y
Sbjct 640 RELNKAFCGFYRPEVNP---QNLSAVATGNWGCGAFGGDPRLKALIQLLAAAEVGRDLVY 696
Query 123 MAFGDKSL 130
FGD+ L
Sbjct 697 FTFGDREL 704
> dre:797792 si:dkey-259k14.2
Length=609
Score = 81.3 bits (199), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 42/106 (39%), Positives = 58/106 (54%), Gaps = 8/106 (7%)
Query 32 DGLGRRAATVAAIDALIVRNPNSQYQENKMKREVLKALLGFQGDPFEALLGEPKAPVATG 91
D RR + AIDAL +NP QY + RE+ KA +GF G+PK +ATG
Sbjct 463 DVWKRRFRQIVAIDALDFKNPLEQYSRENITRELNKAFVGF--------CGQPKTAIATG 514
Query 92 KWGCVIFGGDNQLKTVLQWIAASAAARPLQYMAFGDKSLSGMQQRL 137
WGC F GD +LK +LQ +AA+ R + Y FG+ L+ Q++
Sbjct 515 NWGCGAFRGDPKLKALLQLMAAAVVDRDVAYFTFGNTHLANELQKM 560
> cel:F20C5.1 pme-3; Poly(ADP-ribose) Metabolism Enzyme family
member (pme-3)
Length=764
Score = 69.3 bits (168), Expect = 6e-12, Method: Composition-based stats.
Identities = 40/116 (34%), Positives = 59/116 (50%), Gaps = 13/116 (11%)
Query 32 DGLGRRAATVAAIDALIVRNPNSQYQENKMK-----REVLKALLGF--QGDPFEALLGEP 84
D GR AIDA++ + Q ++ RE+ KA +GF QG F +
Sbjct 600 DRFGRLRVETIAIDAILFKGSKLDCQTEQLNKANIIREMKKASIGFMSQGPKFTNI---- 655
Query 85 KAPVATGKWGCVIFGGDNQLKTVLQWIAASAAARPLQYMAFGDKSLSGMQQRLLRR 140
P+ TG WGC F GD LK ++Q IAA A RPL + +FG+ L+ ++++ R
Sbjct 656 --PIVTGWWGCGAFNGDKPLKFIIQVIAAGVADRPLHFCSFGEPELAAKCKKIIER 709
> cel:H23L24.5 pme-4; Poly(ADP-ribose) Metabolism Enzyme family
member (pme-4); K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=485
Score = 66.2 bits (160), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 51/138 (36%), Positives = 66/138 (47%), Gaps = 20/138 (14%)
Query 3 YRGYERTFELRAAEVPEGCHWSLHGLCPLDGLGRRAATVAAIDALIVRNPNS-------Q 55
Y GY T L+ A++ H + + D GR AIDA VRN + Q
Sbjct 308 YTGYSNT--LKWAKITPK-HSAQNNNSFRDQFGRLQTETVAIDA--VRNAGTPLECLLNQ 362
Query 56 YQENKMKREVLKALLGF--QGDPFEALLGEPKAPVATGKWGCVIFGGDNQLKTVLQWIAA 113
K+ REV KA +GF GD F K PV +G WGC F G+ LK ++Q IA
Sbjct 363 LTTEKLTREVRKAAIGFLSAGDGFS------KIPVVSGWWGCGAFRGNKPLKFLIQVIAC 416
Query 114 SAAARPLQYMAFGDKSLS 131
+ RPLQ+ FGD L+
Sbjct 417 GISDRPLQFCTFGDTELA 434
> mmu:26430 Parg, AI413217; poly (ADP-ribose) glycohydrolase (EC:3.2.1.143);
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=961
Score = 63.5 bits (153), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 45/101 (44%), Positives = 57/101 (56%), Gaps = 7/101 (6%)
Query 32 DGLGRRAATVAAIDALIVRNPNSQYQENKMKREVLKALLGF--QGDPFEALLGEPKAPVA 89
D RR + AIDAL R Q+ K++RE+ KA GF G P E L + VA
Sbjct 805 DDWQRRCTEIVAIDALHFRRYLDQFVPEKVRRELNKAYCGFLRPGVPSENL-----SAVA 859
Query 90 TGKWGCVIFGGDNQLKTVLQWIAASAAARPLQYMAFGDKSL 130
TG WGC FGGD +LK ++Q +AA+AA R + Y FGD L
Sbjct 860 TGNWGCGAFGGDARLKALIQILAAAAAERDVVYFTFGDSEL 900
> ath:AT2G31865 poly (ADP-ribose) glycohydrolase (PARG) family
protein
Length=522
Score = 62.4 bits (150), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 49/143 (34%), Positives = 66/143 (46%), Gaps = 39/143 (27%)
Query 31 LDGLGRRAATVAAIDALIVRNPN-SQYQENKMKREVLKALLGF----------QGDPFEA 79
LD RR V AIDA+ +P QY+ + + REV KA G+ + DP EA
Sbjct 340 LDIFRRRKTRVIAIDAM--PDPGMGQYKLDALIREVNKAFSGYMHQCKYNIDVKHDP-EA 396
Query 80 LLG------------------------EPKAPVATGKWGCVIFGGDNQLKTVLQWIAASA 115
E K VATG WGC +FGGD +LK +LQW+A S
Sbjct 397 SSSHVPLTSDSASQVIESSHRWCIDHEEKKIGVATGNWGCGVFGGDPELKIMLQWLAISQ 456
Query 116 AARP-LQYMAFGDKSLSGMQQRL 137
+ RP + Y FG ++L + Q +
Sbjct 457 SGRPFMSYYTFGLQALQNLNQVI 479
> hsa:8505 PARG, FLJ54459, FLJ60257, FLJ60456, PARG99; poly (ADP-ribose)
glycohydrolase (EC:3.2.1.143); K07759 poly(ADP-ribose)
glycohydrolase [EC:3.2.1.143]
Length=976
Score = 62.0 bits (149), Expect = 9e-10, Method: Composition-based stats.
Identities = 43/99 (43%), Positives = 55/99 (55%), Gaps = 3/99 (3%)
Query 32 DGLGRRAATVAAIDALIVRNPNSQYQENKMKREVLKALLGFQGDPFEALLGEPKAPVATG 91
D RR + AIDAL R Q+ KM+RE+ KA GF + E + VATG
Sbjct 812 DDWQRRCTEIVAIDALHFRRYLDQFVPEKMRRELNKAYCGFLR---PGVSSENLSAVATG 868
Query 92 KWGCVIFGGDNQLKTVLQWIAASAAARPLQYMAFGDKSL 130
WGC FGGD +LK ++Q +AA+AA R + Y FGD L
Sbjct 869 NWGCGAFGGDARLKALIQILAAAAAERDVVYFTFGDSEL 907
> tgo:TGME49_062760 poly(ADP-ribose) glycohydrolase, putative
Length=952
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 35/99 (35%), Positives = 47/99 (47%), Gaps = 3/99 (3%)
Query 49 VRNPNSQYQENKMKREVLKALLGFQGDPFEALLGEPKAPVATGKWGCVIFGGDNQLKTVL 108
V P+ ++N+ + + P + L+ K P ATG WGC +F GD QLK +L
Sbjct 792 VSTPDGALRQNEERDRFSASEATLAPGPHQGLV---KRPFATGNWGCGVFKGDPQLKFLL 848
Query 109 QWIAASAAARPLQYMAFGDKSLSGMQQRLLRRGPSSSTD 147
QW+AAS R L Y A L G R R G + D
Sbjct 849 QWLAASLVGRRLIYHAHSRPELVGSASRSQRGGTVTEQD 887
> ath:AT2G31870 TEJ; TEJ (Sanskrit for 'bright'); poly(ADP-ribose)
glycohydrolase
Length=547
Score = 57.4 bits (137), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 25/49 (51%), Positives = 33/49 (67%), Gaps = 1/49 (2%)
Query 88 VATGKWGCVIFGGDNQLKTVLQWIAASAAARP-LQYMAFGDKSLSGMQQ 135
VATG WGC +FGGD +LK +QW+AAS RP + Y FG ++L + Q
Sbjct 446 VATGNWGCGVFGGDPELKATIQWLAASQTRRPFISYYTFGVEALRNLDQ 494
> hsa:10566 AKAP3, AKAP110, CT82, FSP95, PRKA3, SOB1; A kinase
(PRKA) anchor protein 3
Length=853
Score = 35.4 bits (80), Expect = 0.100, Method: Composition-based stats.
Identities = 17/39 (43%), Positives = 26/39 (66%), Gaps = 3/39 (7%)
Query 101 DNQLKTVLQWIAASAAARPLQYMAFGDKSLSGMQQRLLR 139
+ QL+ VLQW+AAS P+ Y A D+ G+Q++LL+
Sbjct 770 NKQLQAVLQWVAASELNVPILYFAGDDE---GIQEKLLQ 805
> mmu:11642 Akap3, Prka3; A kinase (PRKA) anchor protein 3
Length=864
Score = 35.4 bits (80), Expect = 0.10, Method: Composition-based stats.
Identities = 17/39 (43%), Positives = 26/39 (66%), Gaps = 3/39 (7%)
Query 101 DNQLKTVLQWIAASAAARPLQYMAFGDKSLSGMQQRLLR 139
+ QL+ VLQW+AAS P+ Y A D+ G+Q++LL+
Sbjct 781 NKQLQAVLQWVAASELNVPILYFAGDDE---GIQEKLLQ 816
> cel:F56D2.5 hypothetical protein; K11971 E3 ubiquitin-protein
ligase RNF14 [EC:6.3.2.19]
Length=437
Score = 33.1 bits (74), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 11/44 (25%), Positives = 25/44 (56%), Gaps = 0/44 (0%)
Query 98 FGGDNQLKTVLQWIAASAAARPLQYMAFGDKSLSGMQQRLLRRG 141
F +++ + + +W A A + Y +G+K++ +++R L RG
Sbjct 317 FRKEDEERIMKEWNEADEAGKEEMYKRYGEKNMKALEERFLNRG 360
> tgo:TGME49_054960 hypothetical protein
Length=1465
Score = 30.8 bits (68), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 19/61 (31%), Positives = 24/61 (39%), Gaps = 10/61 (16%)
Query 81 LGEPKAPVATGKWGCVIFGGDNQLKTVLQWIAASAAARPLQYMAFGDKSLSGMQQRLLRR 140
LGEPKAP W +F K+ W++ + P AF D G R R
Sbjct 593 LGEPKAPEEASSWFNTLF------KSPADWLSPDRSRSP----AFADDGAGGADSRSRRH 642
Query 141 G 141
G
Sbjct 643 G 643
> xla:496232 ppp4r2-a; serine/threonine-protein phosphatase 4
regulatory subunit 2-A
Length=403
Score = 29.6 bits (65), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 13/28 (46%), Positives = 19/28 (67%), Gaps = 1/28 (3%)
Query 51 NPNSQYQE-NKMKREVLKALLGFQGDPF 77
NPN +Y ++MK+ +LK + GF G PF
Sbjct 72 NPNVEYIPFDEMKQRILKIVTGFNGTPF 99
> cel:F52F12.4 lsl-1; LSY-2-Like family member (lsl-1)
Length=318
Score = 28.9 bits (63), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 23/42 (54%), Gaps = 2/42 (4%)
Query 35 GRRAATVAAIDALIVRNPNSQYQENKMKREVLKALLGFQGDP 76
RR++ + DAL++R + Y + KR V K LLG DP
Sbjct 132 NRRSSGIIPSDALVIRGTSMPYYNPEKKRSVPKLLLG--KDP 171
Lambda K H
0.321 0.136 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4341553636
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40