bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7477_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
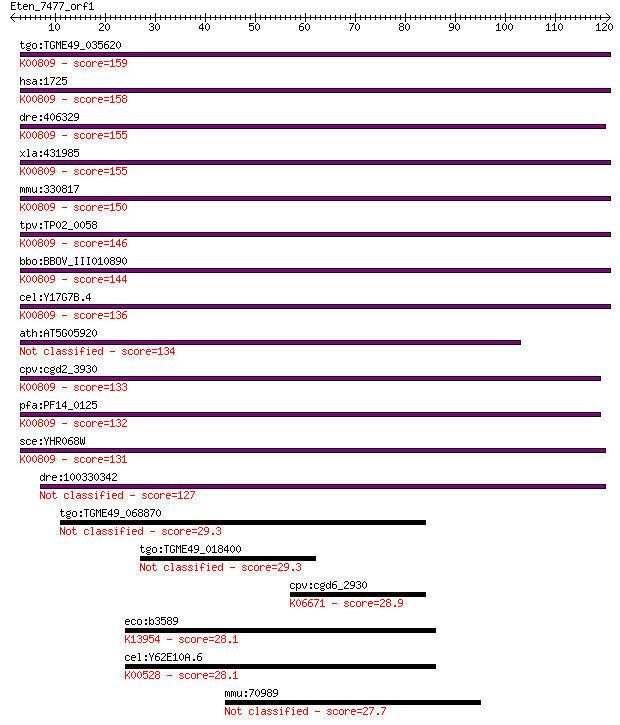
tgo:TGME49_035620 deoxyhypusine synthase, putative (EC:2.5.1.4... 159 1e-39
hsa:1725 DHPS, DHS, MIG13; deoxyhypusine synthase (EC:2.5.1.46... 158 4e-39
dre:406329 dhps, wu:fb52g04, wu:fj48f12, zgc:111900, zgc:56018... 155 2e-38
xla:431985 dhps, MGC82178; deoxyhypusine synthase (EC:2.5.1.46... 155 4e-38
mmu:330817 Dhps, Dhs, MGC49129, MGC74384; deoxyhypusine syntha... 150 8e-37
tpv:TP02_0058 deoxyhypusine synthase; K00809 deoxyhypusine syn... 146 2e-35
bbo:BBOV_III010890 17.m07937; deoxyhypusine synthase; K00809 d... 144 8e-35
cel:Y17G7B.4 hypothetical protein; K00809 deoxyhypusine syntha... 136 2e-32
ath:AT5G05920 DHS; DHS (DEOXYHYPUSINE SYNTHASE) 134 6e-32
cpv:cgd2_3930 deoxyhypusine synthase ; K00809 deoxyhypusine sy... 133 1e-31
pfa:PF14_0125 deoxyhypusine synthase (EC:2.5.1.46); K00809 deo... 132 2e-31
sce:YHR068W DYS1; Deoxyhypusine synthase, catalyzes formation ... 131 6e-31
dre:100330342 deoxyhypusine synthase-like 127 1e-29
tgo:TGME49_068870 TPR domain-containing protein (EC:1.1.99.28) 29.3 2.9
tgo:TGME49_018400 serine/threonine-protein kinase, putative (E... 29.3 3.0
cpv:cgd6_2930 protein with possible stromal antigen (SA/STAG) ... 28.9 4.5
eco:b3589 yiaY, ECK3578, JW5648; predicted Fe-containing alcoh... 28.1 7.1
cel:Y62E10A.6 hypothetical protein; K00528 ferredoxin--NADP+ r... 28.1 7.7
mmu:70989 4931429I11Rik, BB239335; RIKEN cDNA 4931429I11 gene 27.7 8.9
> tgo:TGME49_035620 deoxyhypusine synthase, putative (EC:2.5.1.46);
K00809 deoxyhypusine synthase [EC:2.5.1.46]
Length=457
Score = 159 bits (403), Expect = 1e-39, Method: Composition-based stats.
Identities = 74/118 (62%), Positives = 90/118 (76%), Gaps = 0/118 (0%)
Query 3 SLGDNLYFHHFRSPGLVLDVARDVAAANQLAAAACCSGLLLLGGGAPKHHACNAMLMRGG 62
SLGDNLYFH +R GL++D+ +DV N +A SG+++LGGG PKHH CNA LMR G
Sbjct 331 SLGDNLYFHSYRHKGLIVDIVQDVRNINDIATKCKKSGMIILGGGLPKHHTCNANLMRNG 390
Query 63 ADFAVFLNSASDFDGSDSGARPDEAKSWGKLRAGALTAKVTADASLTFPLLVAATFAK 120
ADFAV++NS S+FDGSDSGARPDEA SWGK+ A KV DA++ FPL+VAATFAK
Sbjct 391 ADFAVYINSGSEFDGSDSGARPDEAVSWGKITCEAKPVKVHGDATVLFPLVVAATFAK 448
> hsa:1725 DHPS, DHS, MIG13; deoxyhypusine synthase (EC:2.5.1.46);
K00809 deoxyhypusine synthase [EC:2.5.1.46]
Length=369
Score = 158 bits (399), Expect = 4e-39, Method: Composition-based stats.
Identities = 72/118 (61%), Positives = 90/118 (76%), Gaps = 0/118 (0%)
Query 3 SLGDNLYFHHFRSPGLVLDVARDVAAANQLAAAACCSGLLLLGGGAPKHHACNAMLMRGG 62
SLGD ++FH +++PGLVLD+ D+ N A A C+G+++LGGG KHH NA LMR G
Sbjct 240 SLGDMIFFHSYKNPGLVLDIVEDLRLINTQAIFAKCTGMIILGGGVVKHHIANANLMRNG 299
Query 63 ADFAVFLNSASDFDGSDSGARPDEAKSWGKLRAGALTAKVTADASLTFPLLVAATFAK 120
AD+AV++N+A +FDGSDSGARPDEA SWGK+R A KV ADASL FPLLVA TFA+
Sbjct 300 ADYAVYINTAQEFDGSDSGARPDEAVSWGKIRVDAQPVKVYADASLVFPLLVAETFAQ 357
> dre:406329 dhps, wu:fb52g04, wu:fj48f12, zgc:111900, zgc:56018,
zgc:77325; deoxyhypusine synthase (EC:2.5.1.46); K00809
deoxyhypusine synthase [EC:2.5.1.46]
Length=361
Score = 155 bits (393), Expect = 2e-38, Method: Composition-based stats.
Identities = 73/117 (62%), Positives = 87/117 (74%), Gaps = 0/117 (0%)
Query 3 SLGDNLYFHHFRSPGLVLDVARDVAAANQLAAAACCSGLLLLGGGAPKHHACNAMLMRGG 62
SLGD +YFH +++PGLVLD+ D+ N A A +G+++LGGG KHH NA LMR G
Sbjct 232 SLGDMIYFHSYKNPGLVLDIVEDIRRLNSKAVFAKSTGMIILGGGLVKHHIANANLMRNG 291
Query 63 ADFAVFLNSASDFDGSDSGARPDEAKSWGKLRAGALTAKVTADASLTFPLLVAATFA 119
ADFAVF+N+ +FDGSDSGARPDEA SWGK+R A KV ADASL FPLLVA TFA
Sbjct 292 ADFAVFVNTGQEFDGSDSGARPDEAISWGKIRMDASPVKVYADASLVFPLLVAETFA 348
> xla:431985 dhps, MGC82178; deoxyhypusine synthase (EC:2.5.1.46);
K00809 deoxyhypusine synthase [EC:2.5.1.46]
Length=349
Score = 155 bits (391), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 71/118 (60%), Positives = 88/118 (74%), Gaps = 0/118 (0%)
Query 3 SLGDNLYFHHFRSPGLVLDVARDVAAANQLAAAACCSGLLLLGGGAPKHHACNAMLMRGG 62
SLGD LYFH +++PGL LD+ D+ N A +G+++LGGG KHH CNA LMR G
Sbjct 219 SLGDMLYFHSYKNPGLKLDIVEDIRKLNSQVIFAKKTGMIILGGGLIKHHICNANLMRNG 278
Query 63 ADFAVFLNSASDFDGSDSGARPDEAKSWGKLRAGALTAKVTADASLTFPLLVAATFAK 120
AD+AVF+N+A +FDGSD+GARPDEA SWGK+R A K+ ADASL FPLLVA TFA+
Sbjct 279 ADYAVFVNTAQEFDGSDAGARPDEAVSWGKIRMEAKPVKIYADASLVFPLLVAQTFAQ 336
> mmu:330817 Dhps, Dhs, MGC49129, MGC74384; deoxyhypusine synthase
(EC:2.5.1.46); K00809 deoxyhypusine synthase [EC:2.5.1.46]
Length=369
Score = 150 bits (379), Expect = 8e-37, Method: Composition-based stats.
Identities = 72/118 (61%), Positives = 89/118 (75%), Gaps = 0/118 (0%)
Query 3 SLGDNLYFHHFRSPGLVLDVARDVAAANQLAAAACCSGLLLLGGGAPKHHACNAMLMRGG 62
SLGD ++FH +++PGLVLD+ D+ N A A SG+++LGGG KHH NA LMR G
Sbjct 240 SLGDMIFFHSYKNPGLVLDIVEDLRLINTQAIFAKRSGMIILGGGVVKHHIANANLMRNG 299
Query 63 ADFAVFLNSASDFDGSDSGARPDEAKSWGKLRAGALTAKVTADASLTFPLLVAATFAK 120
AD+AV++N+A +FDGSDSGARPDEA SWGK+R A KV ADASL FPLLVA TFA+
Sbjct 300 ADYAVYINTAQEFDGSDSGARPDEAVSWGKIRMDAQPVKVYADASLVFPLLVAETFAQ 357
> tpv:TP02_0058 deoxyhypusine synthase; K00809 deoxyhypusine synthase
[EC:2.5.1.46]
Length=370
Score = 146 bits (368), Expect = 2e-35, Method: Composition-based stats.
Identities = 69/121 (57%), Positives = 87/121 (71%), Gaps = 3/121 (2%)
Query 3 SLGDNLYFHHFR--SPG-LVLDVARDVAAANQLAAAACCSGLLLLGGGAPKHHACNAMLM 59
SLGDNLYFH +R SP L LD+ +D+ A N LA SGL++LGGG PKHH CN+ LM
Sbjct 243 SLGDNLYFHTYRKSSPTTLYLDIVKDIRAINDLAVRCKKSGLIILGGGLPKHHVCNSNLM 302
Query 60 RGGADFAVFLNSASDFDGSDSGARPDEAKSWGKLRAGALTAKVTADASLTFPLLVAATFA 119
R GADFA+++++A ++DGSDSGA PDEA SWGK++ KV ADAS+ FPL+VA
Sbjct 303 RNGADFAIYISTAQEYDGSDSGANPDEAVSWGKIKPNTDPVKVHADASIVFPLIVAGVLK 362
Query 120 K 120
K
Sbjct 363 K 363
> bbo:BBOV_III010890 17.m07937; deoxyhypusine synthase; K00809
deoxyhypusine synthase [EC:2.5.1.46]
Length=370
Score = 144 bits (362), Expect = 8e-35, Method: Composition-based stats.
Identities = 71/121 (58%), Positives = 87/121 (71%), Gaps = 3/121 (2%)
Query 3 SLGDNLYFHHFR--SPG-LVLDVARDVAAANQLAAAACCSGLLLLGGGAPKHHACNAMLM 59
S+GDNLYFH + SP L +DV RD+ N +A + SG+++LGGG KHH CNA LM
Sbjct 244 SIGDNLYFHTYSKSSPTTLKIDVVRDIRRVNDIAVHSKKSGIIILGGGLAKHHVCNANLM 303
Query 60 RGGADFAVFLNSASDFDGSDSGARPDEAKSWGKLRAGALTAKVTADASLTFPLLVAATFA 119
R GADFAV++++A +FDGSDSGA PDEA SWGK++A KV ADASL FPLLVA F
Sbjct 304 RNGADFAVYISTAQEFDGSDSGANPDEAVSWGKIKAYTNPVKVHADASLVFPLLVAGVFR 363
Query 120 K 120
K
Sbjct 364 K 364
> cel:Y17G7B.4 hypothetical protein; K00809 deoxyhypusine synthase
[EC:2.5.1.46]
Length=371
Score = 136 bits (342), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 67/119 (56%), Positives = 84/119 (70%), Gaps = 1/119 (0%)
Query 3 SLGDNLYFHHFRS-PGLVLDVARDVAAANQLAAAACCSGLLLLGGGAPKHHACNAMLMRG 61
SLGD LYFH +S PGL +D+ DV N +A + +G ++LGGG KHH NA LMR
Sbjct 247 SLGDMLYFHSVKSSPGLRVDIVEDVRHINTIAVKSFKTGSIILGGGVVKHHINNANLMRN 306
Query 62 GADFAVFLNSASDFDGSDSGARPDEAKSWGKLRAGALTAKVTADASLTFPLLVAATFAK 120
GAD V++N+ +FDGSDSGA+PDEA SWGK++ A KV A+A+L FPLLVA TFAK
Sbjct 307 GADHTVYINTGQEFDGSDSGAQPDEAVSWGKVKPSAGAVKVHAEATLVFPLLVAETFAK 365
> ath:AT5G05920 DHS; DHS (DEOXYHYPUSINE SYNTHASE)
Length=356
Score = 134 bits (337), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 62/102 (60%), Positives = 77/102 (75%), Gaps = 2/102 (1%)
Query 3 SLGDNLYFHHFRSPGLVLDVARDVAAANQLAAAACC--SGLLLLGGGAPKHHACNAMLMR 60
SLGD LYFH FR+ GL++DV +D+ A N A A +G+++LGGG PKHH CNA +MR
Sbjct 239 SLGDMLYFHSFRTSGLIIDVVQDIRAMNGEAVHANPKKTGMIILGGGLPKHHICNANMMR 298
Query 61 GGADFAVFLNSASDFDGSDSGARPDEAKSWGKLRAGALTAKV 102
GAD+AVF+N+ +FDGSDSGARPDEA SWGK+R A T KV
Sbjct 299 NGADYAVFINTGQEFDGSDSGARPDEAVSWGKIRGSAKTVKV 340
> cpv:cgd2_3930 deoxyhypusine synthase ; K00809 deoxyhypusine
synthase [EC:2.5.1.46]
Length=362
Score = 133 bits (335), Expect = 1e-31, Method: Composition-based stats.
Identities = 66/116 (56%), Positives = 88/116 (75%), Gaps = 0/116 (0%)
Query 3 SLGDNLYFHHFRSPGLVLDVARDVAAANQLAAAACCSGLLLLGGGAPKHHACNAMLMRGG 62
SLGDNL+FH +R+PGL +DV D+ N +A SG+++LGGG PKHH CNA LMR G
Sbjct 244 SLGDNLFFHSYRNPGLKVDVVEDIRRINDIALNIKKSGIIILGGGIPKHHTCNANLMRNG 303
Query 63 ADFAVFLNSASDFDGSDSGARPDEAKSWGKLRAGALTAKVTADASLTFPLLVAATF 118
AD+AV+++SA++FDGSDSGA PDEA SWGK++A + KV DA++ FPL+ +TF
Sbjct 304 ADYAVYISSATEFDGSDSGASPDEAVSWGKIKAESTPVKVFGDATIIFPLIFYSTF 359
> pfa:PF14_0125 deoxyhypusine synthase (EC:2.5.1.46); K00809 deoxyhypusine
synthase [EC:2.5.1.46]
Length=496
Score = 132 bits (333), Expect = 2e-31, Method: Composition-based stats.
Identities = 63/121 (52%), Positives = 84/121 (69%), Gaps = 5/121 (4%)
Query 3 SLGDNLYFHHFRSP---GLVLDVARDVAAANQLAAAACCSGLLLLGGGAPKHHACNAMLM 59
SLGDNL+FH++ L+LD+ +D+ N LA SG+++LGGG PKHH CNA LM
Sbjct 365 SLGDNLFFHNYGKKIKNNLILDIVKDIKKINSLAMNCEKSGIIILGGGLPKHHVCNANLM 424
Query 60 RGGADFAVFLNSASDFDGSDSGARPDEAKSWGKLRAGALT--AKVTADASLTFPLLVAAT 117
R GADFAV++N+AS++DGSDSGA EA SWGK++ G KV DA++ FPL+V +
Sbjct 425 RNGADFAVYVNTASEYDGSDSGANTTEALSWGKIKYGQTNNHVKVFGDATILFPLMVLNS 484
Query 118 F 118
F
Sbjct 485 F 485
> sce:YHR068W DYS1; Deoxyhypusine synthase, catalyzes formation
of deoxyhypusine, the first step in hypusine biosynthesis;
triggers posttranslational hypusination of translation elongation
factor eIF-5A and regulates its intracellular levels;
tetrameric (EC:2.5.1.46); K00809 deoxyhypusine synthase [EC:2.5.1.46]
Length=387
Score = 131 bits (329), Expect = 6e-31, Method: Composition-based stats.
Identities = 62/119 (52%), Positives = 87/119 (73%), Gaps = 2/119 (1%)
Query 3 SLGDNLYFHHFR-SPG-LVLDVARDVAAANQLAAAACCSGLLLLGGGAPKHHACNAMLMR 60
S+GD L+FH F+ SP L +D+ D+ N ++ AA +G+++LGGG KHH NA LMR
Sbjct 259 SIGDMLFFHTFKASPKQLRVDIVGDIRKINSMSMAAYRAGMIILGGGLIKHHIANACLMR 318
Query 61 GGADFAVFLNSASDFDGSDSGARPDEAKSWGKLRAGALTAKVTADASLTFPLLVAATFA 119
GAD+AV++N+ ++DGSD+GARPDEA SWGK++A A + K+ AD + PL+VAATFA
Sbjct 319 NGADYAVYINTGQEYDGSDAGARPDEAVSWGKIKAEAKSVKLFADVTTVLPLIVAATFA 377
> dre:100330342 deoxyhypusine synthase-like
Length=312
Score = 127 bits (318), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 66/113 (58%), Positives = 76/113 (67%), Gaps = 4/113 (3%)
Query 7 NLYFHHFRSPGLVLDVARDVAAANQLAAAACCSGLLLLGGGAPKHHACNAMLMRGGADFA 66
NL FHH + D+ R N A A +G+++LGGG KHH NA LMR GADFA
Sbjct 191 NLDFHHDNISPPISDIRR----LNSKAVFAKSTGMIILGGGLVKHHIANANLMRNGADFA 246
Query 67 VFLNSASDFDGSDSGARPDEAKSWGKLRAGALTAKVTADASLTFPLLVAATFA 119
VF+N+ +FDGSDSGARPDEA SWGK+R A KV ADASL FPLLVA TFA
Sbjct 247 VFVNTGQEFDGSDSGARPDEAISWGKIRMDASPVKVYADASLVFPLLVAETFA 299
> tgo:TGME49_068870 TPR domain-containing protein (EC:1.1.99.28)
Length=1031
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 29/77 (37%), Positives = 32/77 (41%), Gaps = 25/77 (32%)
Query 11 HHFRSPGL----VLDVARDVAAANQLAAAACCSGLLLLGGGAPKHHACNAMLMRGGADFA 66
H FR GL L V DVA AN A CS KH A + +G DFA
Sbjct 170 HLFRDLGLPEKPFLYVFSDVAEAN----VAFCS----------KHPAFQEFIQQGWLDFA 215
Query 67 VFLNSASDFDGSDSGAR 83
V FDG+D AR
Sbjct 216 V-------FDGNDKEAR 225
> tgo:TGME49_018400 serine/threonine-protein kinase, putative
(EC:2.7.11.1)
Length=2911
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 18/35 (51%), Gaps = 0/35 (0%)
Query 27 AAANQLAAAACCSGLLLLGGGAPKHHACNAMLMRG 61
A + A ACC+G L G G P + C A +RG
Sbjct 1864 ATEMRRARKACCTGRLHAGKGDPGNFECRAWSLRG 1898
> cpv:cgd6_2930 protein with possible stromal antigen (SA/STAG)
domain ; K06671 cohesin complex subunit SA-1/2
Length=1404
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 12/27 (44%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 57 MLMRGGADFAVFLNSASDFDGSDSGAR 83
+++ GG +VFL S SD+ GSDS +
Sbjct 108 LILEGGGIPSVFLQSISDWTGSDSAEK 134
> eco:b3589 yiaY, ECK3578, JW5648; predicted Fe-containing alcohol
dehydrogenase, Pfam00465 family; K13954 alcohol dehydrogenase
[EC:1.1.1.1]
Length=383
Score = 28.1 bits (61), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 30/65 (46%), Gaps = 12/65 (18%)
Query 24 RDVAAANQLAAAACCSGLLLLGGGAPKHHACN---AMLMRGGADFAVFLNSASDFDGSDS 80
+VAA +L C ++ LGGG+P H C A++ G D D++G D
Sbjct 75 ENVAAGLKLLKENNCDSVISLGGGSP--HDCAKGIALVAANGGDI-------RDYEGVDR 125
Query 81 GARPD 85
A+P
Sbjct 126 SAKPQ 130
> cel:Y62E10A.6 hypothetical protein; K00528 ferredoxin--NADP+
reductase [EC:1.18.1.2]
Length=458
Score = 28.1 bits (61), Expect = 7.7, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 27/65 (41%), Gaps = 3/65 (4%)
Query 24 RDVAAANQLAAAACCSG---LLLLGGGAPKHHACNAMLMRGGADFAVFLNSASDFDGSDS 80
R +A +N + SG L ++G G ACN +L + VF NS F
Sbjct 8 RRIACSNLNLKSRKLSGKPRLAIVGSGPAGMFACNGLLRKSNFSVDVFENSPVPFGLVRY 67
Query 81 GARPD 85
G PD
Sbjct 68 GVAPD 72
> mmu:70989 4931429I11Rik, BB239335; RIKEN cDNA 4931429I11 gene
Length=770
Score = 27.7 bits (60), Expect = 8.9, Method: Composition-based stats.
Identities = 21/54 (38%), Positives = 26/54 (48%), Gaps = 4/54 (7%)
Query 44 LGGGAPKHHACNAMLMRGGADFAV--FLNSAS-DFDGSDSGARPDEAKSWGKLR 94
L +P HH N + G F FLN S D SDS + P EAKSW ++
Sbjct 10 LSSQSPVHHT-NLRIPPSGRPFKKEDFLNLISKDSLESDSESLPQEAKSWSDIK 62
Lambda K H
0.320 0.133 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2018002440
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40