bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
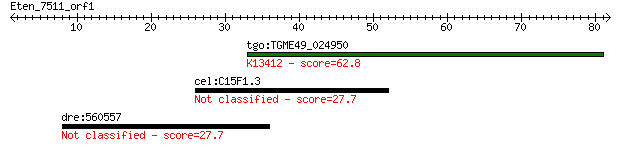
Query= Eten_7511_orf1
Length=81
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_024950 protein kinase 6, putative (EC:1.6.3.1 2.7.1... 62.8 2e-10
cel:C15F1.3 tra-2; TRAnsformer : XX animals transformed into m... 27.7 8.4
dre:560557 SH2 domain containing 4B-like 27.7 9.0
> tgo:TGME49_024950 protein kinase 6, putative (EC:1.6.3.1 2.7.11.17);
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=681
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 32/56 (57%), Positives = 38/56 (67%), Gaps = 8/56 (14%)
Query 33 ADHHCHGPE-----EKPHFFSLCTKCATDGKDKPL---REPAKGFITRSESYVNAS 80
AD+ C G + KPHFFSLCTKCA +GKDK REP +G I RSES++NAS
Sbjct 92 ADYGCRGGDCCDTSTKPHFFSLCTKCANEGKDKSQHDRREPVRGLIIRSESFMNAS 147
> cel:C15F1.3 tra-2; TRAnsformer : XX animals transformed into
males family member (tra-2)
Length=1475
Score = 27.7 bits (60), Expect = 8.4, Method: Composition-based stats.
Identities = 11/27 (40%), Positives = 16/27 (59%), Gaps = 1/27 (3%)
Query 26 PCRPNCAAD-HHCHGPEEKPHFFSLCT 51
P + CAA H C P + H+F++CT
Sbjct 238 PRQKTCAASIHSCDTPLDSEHYFNICT 264
> dre:560557 SH2 domain containing 4B-like
Length=423
Score = 27.7 bits (60), Expect = 9.0, Method: Composition-based stats.
Identities = 13/29 (44%), Positives = 19/29 (65%), Gaps = 1/29 (3%)
Query 8 NMQKDKRLDTAEGRPLESPCRP-NCAADH 35
N K++ + T+ G L+ PCRP + AADH
Sbjct 390 NFHKEEVITTSGGELLQDPCRPRSSAADH 418
Lambda K H
0.314 0.131 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2053886800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40