bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7515_orf1
Length=83
Score E
Sequences producing significant alignments: (Bits) Value
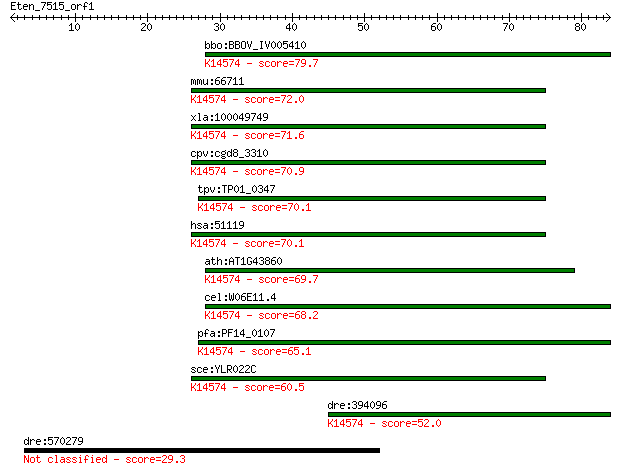
bbo:BBOV_IV005410 23.m05967; hypothetical protein; K14574 ribo... 79.7 2e-15
mmu:66711 Sbds, 4733401P19Rik, AI836084, CGI-97, SDS; Shwachma... 72.0 4e-13
xla:100049749 sbds; Shwachman-Bodian-Diamond syndrome; K14574 ... 71.6 6e-13
cpv:cgd8_3310 S. cerevisiae Ylr022cp like protein that has a C... 70.9 9e-13
tpv:TP01_0347 hypothetical protein; K14574 ribosome maturation... 70.1 2e-12
hsa:51119 SBDS, FLJ10917, SDS, SWDS; Shwachman-Bodian-Diamond ... 70.1 2e-12
ath:AT1G43860 transcription factor; K14574 ribosome maturation... 69.7 2e-12
cel:W06E11.4 sbds-1; Shwachman-Bodian-Diamond Syndrome protein... 68.2 7e-12
pfa:PF14_0107 conserved Plasmodium protein, unknown function; ... 65.1 6e-11
sce:YLR022C SDO1; Essential protein involved in 60S ribosome m... 60.5 1e-09
dre:394096 sbds, MGC56700, zgc:56700; Shwachman-Bodian-Diamond... 52.0 5e-07
dre:570279 zgc:194273; zgc:194281 29.3 3.0
> bbo:BBOV_IV005410 23.m05967; hypothetical protein; K14574 ribosome
maturation protein SDO1
Length=393
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 37/56 (66%), Positives = 44/56 (78%), Gaps = 5/56 (8%)
Query 28 LKQPVTQVRLTNVAVVRLRLGGERFEVACYKNKVVNWREGKETDLNEASSSSVMQT 83
L QP+ QVRLTNVA VRL+ GG++FE+ACYKNKVVNWR G ETD+ E V+QT
Sbjct 4 LFQPIGQVRLTNVATVRLKTGGKKFEIACYKNKVVNWRSGVETDIQE-----VLQT 54
> mmu:66711 Sbds, 4733401P19Rik, AI836084, CGI-97, SDS; Shwachman-Bodian-Diamond
syndrome homolog (human); K14574 ribosome
maturation protein SDO1
Length=250
Score = 72.0 bits (175), Expect = 4e-13, Method: Composition-based stats.
Identities = 32/49 (65%), Positives = 40/49 (81%), Gaps = 0/49 (0%)
Query 26 MALKQPVTQVRLTNVAVVRLRLGGERFEVACYKNKVVNWREGKETDLNE 74
M++ P Q+RLTNVAVVR++ GG+RFE+ACYKNKVV WR G E DL+E
Sbjct 1 MSIFTPTNQIRLTNVAVVRMKRGGKRFEIACYKNKVVGWRSGVEKDLDE 49
> xla:100049749 sbds; Shwachman-Bodian-Diamond syndrome; K14574
ribosome maturation protein SDO1
Length=250
Score = 71.6 bits (174), Expect = 6e-13, Method: Composition-based stats.
Identities = 31/49 (63%), Positives = 41/49 (83%), Gaps = 0/49 (0%)
Query 26 MALKQPVTQVRLTNVAVVRLRLGGERFEVACYKNKVVNWREGKETDLNE 74
M++ P Q+RLTNVAVVR++ GG+RFE+ACYKNKV++WR G E DL+E
Sbjct 1 MSIFTPTNQIRLTNVAVVRMKKGGKRFEIACYKNKVMSWRSGAEKDLDE 49
> cpv:cgd8_3310 S. cerevisiae Ylr022cp like protein that has a
C2H2 zinc finger and is a component of the exosome ; K14574
ribosome maturation protein SDO1
Length=379
Score = 70.9 bits (172), Expect = 9e-13, Method: Composition-based stats.
Identities = 32/49 (65%), Positives = 40/49 (81%), Gaps = 0/49 (0%)
Query 26 MALKQPVTQVRLTNVAVVRLRLGGERFEVACYKNKVVNWREGKETDLNE 74
M+L QP Q++LTNVAVVR + G+RFEVACYKNK++NWR G E DL+E
Sbjct 7 MSLFQPSNQIKLTNVAVVRYKSHGKRFEVACYKNKILNWRSGVEWDLDE 55
> tpv:TP01_0347 hypothetical protein; K14574 ribosome maturation
protein SDO1
Length=391
Score = 70.1 bits (170), Expect = 2e-12, Method: Composition-based stats.
Identities = 31/48 (64%), Positives = 37/48 (77%), Gaps = 0/48 (0%)
Query 27 ALKQPVTQVRLTNVAVVRLRLGGERFEVACYKNKVVNWREGKETDLNE 74
L QP QVR TNVA+VRL++ GERFE+ACYKNKV NW+ G E D+ E
Sbjct 3 GLFQPTCQVRFTNVAIVRLKVKGERFEIACYKNKVFNWKSGIENDIEE 50
> hsa:51119 SBDS, FLJ10917, SDS, SWDS; Shwachman-Bodian-Diamond
syndrome; K14574 ribosome maturation protein SDO1
Length=250
Score = 70.1 bits (170), Expect = 2e-12, Method: Composition-based stats.
Identities = 31/49 (63%), Positives = 39/49 (79%), Gaps = 0/49 (0%)
Query 26 MALKQPVTQVRLTNVAVVRLRLGGERFEVACYKNKVVNWREGKETDLNE 74
M++ P Q+RLTNVAVVR++ G+RFE+ACYKNKVV WR G E DL+E
Sbjct 1 MSIFTPTNQIRLTNVAVVRMKRAGKRFEIACYKNKVVGWRSGVEKDLDE 49
> ath:AT1G43860 transcription factor; K14574 ribosome maturation
protein SDO1
Length=370
Score = 69.7 bits (169), Expect = 2e-12, Method: Composition-based stats.
Identities = 33/51 (64%), Positives = 39/51 (76%), Gaps = 0/51 (0%)
Query 28 LKQPVTQVRLTNVAVVRLRLGGERFEVACYKNKVVNWREGKETDLNEASSS 78
L QPV Q RLTNVAVVRL+ G RFE+ACYKNKV++WR G E D++E S
Sbjct 5 LVQPVGQKRLTNVAVVRLKKQGNRFEIACYKNKVLSWRSGVEKDIDEVLQS 55
> cel:W06E11.4 sbds-1; Shwachman-Bodian-Diamond Syndrome protein
homolog family member (sbds-1); K14574 ribosome maturation
protein SDO1
Length=253
Score = 68.2 bits (165), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 32/56 (57%), Positives = 40/56 (71%), Gaps = 5/56 (8%)
Query 28 LKQPVTQVRLTNVAVVRLRLGGERFEVACYKNKVVNWREGKETDLNEASSSSVMQT 83
+K P Q LTNVAVVR++ G+RFE+ACYKNKVVNWR E D++E V+QT
Sbjct 5 IKTPTNQKVLTNVAVVRMKKTGKRFEIACYKNKVVNWRNKSEKDIDE-----VLQT 55
> pfa:PF14_0107 conserved Plasmodium protein, unknown function;
K14574 ribosome maturation protein SDO1
Length=638
Score = 65.1 bits (157), Expect = 6e-11, Method: Composition-based stats.
Identities = 25/57 (43%), Positives = 43/57 (75%), Gaps = 0/57 (0%)
Query 27 ALKQPVTQVRLTNVAVVRLRLGGERFEVACYKNKVVNWREGKETDLNEASSSSVMQT 83
AL QP+ QV+ TNVA+V+ + G++FE+ACYKNK+++W+ G E ++++ S ++ T
Sbjct 4 ALFQPINQVKFTNVAIVKYKYKGKKFEIACYKNKIIDWKNGTELNIDDVLQSHLIFT 60
> sce:YLR022C SDO1; Essential protein involved in 60S ribosome
maturation; ortholog of the human protein (SBDS) responsible
for autosomal recessive Shwachman-Bodian-Diamond Syndrome;
highly conserved across archae and eukaryotes; K14574 ribosome
maturation protein SDO1
Length=250
Score = 60.5 bits (145), Expect = 1e-09, Method: Composition-based stats.
Identities = 27/49 (55%), Positives = 39/49 (79%), Gaps = 0/49 (0%)
Query 26 MALKQPVTQVRLTNVAVVRLRLGGERFEVACYKNKVVNWREGKETDLNE 74
M + QP Q++LTNV++VRL+ +RFEVACY+NKV ++R+G E DL+E
Sbjct 1 MPINQPSGQIKLTNVSLVRLKKARKRFEVACYQNKVQDYRKGIEKDLDE 49
> dre:394096 sbds, MGC56700, zgc:56700; Shwachman-Bodian-Diamond
syndrome; K14574 ribosome maturation protein SDO1
Length=231
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 22/39 (56%), Positives = 30/39 (76%), Gaps = 5/39 (12%)
Query 45 LRLGGERFEVACYKNKVVNWREGKETDLNEASSSSVMQT 83
++ GG+RFE+ACYKNKV++WR G E DL+E V+QT
Sbjct 1 MKKGGKRFEIACYKNKVMSWRSGAEKDLDE-----VLQT 34
> dre:570279 zgc:194273; zgc:194281
Length=206
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 26/49 (53%), Gaps = 1/49 (2%)
Query 3 LPVSALRGTSVKTAEVETLTLGNMALKQPVTQVRLTNVAVVRLRLGGER 51
LP+ L+G V + ++ L L M+L++PV + + L L GER
Sbjct 152 LPLCLLKGKRVYSKSLDYLNLDKMSLREPV-DTEVLQYQLQHLNLRGER 199
Lambda K H
0.313 0.126 0.342
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2044675024
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40