bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7540_orf1
Length=83
Score E
Sequences producing significant alignments: (Bits) Value
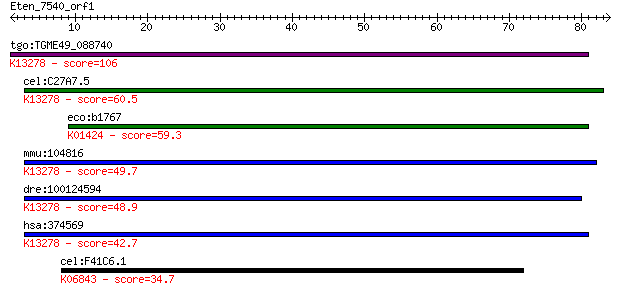
tgo:TGME49_088740 asparaginase, putative (EC:3.1.1.47); K13278... 106 2e-23
cel:C27A7.5 hypothetical protein; K13278 60kDa lysophospholipa... 60.5 1e-09
eco:b1767 ansA, ECK1765, JW1756; cytoplasmic L-asparaginase I ... 59.3 3e-09
mmu:104816 Aspg, A530050D06Rik, AI429460, Gm690; asparaginase ... 49.7 2e-06
dre:100124594 aspg, zgc:171644; asparaginase homolog (S. cerev... 48.9 4e-06
hsa:374569 ASPG, C14orf76; asparaginase homolog (S. cerevisiae... 42.7 3e-04
cel:F41C6.1 unc-6; UNCoordinated family member (unc-6); K06843... 34.7 0.074
> tgo:TGME49_088740 asparaginase, putative (EC:3.1.1.47); K13278
60kDa lysophospholipase [EC:3.1.1.5 3.1.1.47 3.5.1.1]
Length=703
Score = 106 bits (264), Expect = 2e-23, Method: Composition-based stats.
Identities = 50/80 (62%), Positives = 65/80 (81%), Gaps = 0/80 (0%)
Query 1 STRAAIAAGVAVVAATQCHKGTANLLIYENGIWISSLGVICAKDMTPEAVVAKLYYLMGK 60
+ +AAI G+ +VA TQC +G+ANLL YE+G+W+S+LGVI KD+T EA VAKL YLMGK
Sbjct 555 TIKAAIENGINIVATTQCRRGSANLLAYESGVWLSNLGVINGKDLTLEACVAKLAYLMGK 614
Query 61 GVTGRRLKELMESNLRGEVT 80
G TG LK+LMES++RGE+T
Sbjct 615 GYTGAPLKDLMESDIRGELT 634
> cel:C27A7.5 hypothetical protein; K13278 60kDa lysophospholipase
[EC:3.1.1.5 3.1.1.47 3.5.1.1]
Length=713
Score = 60.5 bits (145), Expect = 1e-09, Method: Composition-based stats.
Identities = 35/81 (43%), Positives = 50/81 (61%), Gaps = 2/81 (2%)
Query 3 RAAIAAGVAVVAATQCHKGTANLLIYENGIWISSLGVICAKDMTPEAVVAKLYYLMGKGV 62
+ AIA GV VV +QC KG ++ Y G + +GVI DMT EA +AKL Y++GK
Sbjct 400 KEAIARGVMVVNCSQCLKGQVDV-NYATGKILYDIGVIPGSDMTSEAAMAKLCYVLGKDE 458
Query 63 TGRRLKE-LMESNLRGEVTVG 82
+K +++SNLRGE+T+
Sbjct 459 WDLPMKRSMLQSNLRGEMTIA 479
> eco:b1767 ansA, ECK1765, JW1756; cytoplasmic L-asparaginase
I (EC:3.5.1.1); K01424 L-asparaginase [EC:3.5.1.1]
Length=338
Score = 59.3 bits (142), Expect = 3e-09, Method: Composition-based stats.
Identities = 29/72 (40%), Positives = 42/72 (58%), Gaps = 0/72 (0%)
Query 9 GVAVVAATQCHKGTANLLIYENGIWISSLGVICAKDMTPEAVVAKLYYLMGKGVTGRRLK 68
G+ VV TQC G N+ Y G ++ GVI DMT EA + KL+YL+ + + ++
Sbjct 264 GIVVVNLTQCMSGKVNMGGYATGNALAHAGVIGGADMTVEATLTKLHYLLSQELDTETIR 323
Query 69 ELMESNLRGEVT 80
+ M NLRGE+T
Sbjct 324 KAMSQNLRGELT 335
> mmu:104816 Aspg, A530050D06Rik, AI429460, Gm690; asparaginase
homolog (S. cerevisiae) (EC:3.1.1.5 3.1.1.47 3.5.1.1); K13278
60kDa lysophospholipase [EC:3.1.1.5 3.1.1.47 3.5.1.1]
Length=564
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 29/80 (36%), Positives = 49/80 (61%), Gaps = 2/80 (2%)
Query 3 RAAIAAGVAVVAATQCHKGTANLLIYENGIWISSLGVICAKDMTPEAVVAKLYYLMGK-G 61
R A G+ +V T C +G A Y +G+ ++ G++ DMT EA +AKL Y++G+ G
Sbjct 284 RVAAEQGLIIVNCTHCLQG-AVTSDYASGMAMAGAGIVSGFDMTSEAALAKLSYVLGQPG 342
Query 62 VTGRRLKELMESNLRGEVTV 81
++ K+L+ +LRGE+T+
Sbjct 343 LSLNDRKKLLAKDLRGEMTL 362
> dre:100124594 aspg, zgc:171644; asparaginase homolog (S. cerevisiae);
K13278 60kDa lysophospholipase [EC:3.1.1.5 3.1.1.47
3.5.1.1]
Length=626
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 27/78 (34%), Positives = 45/78 (57%), Gaps = 2/78 (2%)
Query 3 RAAIAAGVAVVAATQCHKGTANLLIYENGIWISSLGVICAKDMTPEAVVAKLYYLMGK-G 61
R A G+ ++ TQC +G+ Y G +S G++ DMTPEA ++KL Y++ K
Sbjct 336 RKATQRGLIMINCTQCLRGSVTT-SYATGQALSDAGLVAGCDMTPEAALSKLSYVLAKQN 394
Query 62 VTGRRLKELMESNLRGEV 79
++ K+++ NLRGE+
Sbjct 395 LSIEEKKKMLSRNLRGEM 412
> hsa:374569 ASPG, C14orf76; asparaginase homolog (S. cerevisiae)
(EC:3.1.1.5 3.1.1.47 3.5.1.1); K13278 60kDa lysophospholipase
[EC:3.1.1.5 3.1.1.47 3.5.1.1]
Length=573
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 32/79 (40%), Positives = 47/79 (59%), Gaps = 2/79 (2%)
Query 3 RAAIAAGVAVVAATQCHKGTANLLIYENGIWISSLGVICAKDMTPEAVVAKLYYLMGK-G 61
R A G+ +V T C +G A Y G+ ++ GVI DMT EA +AKL Y++G+ G
Sbjct 284 RVATERGLVIVNCTHCLQG-AVTTDYAAGMAMAGAGVISGFDMTSEAALAKLSYVLGQPG 342
Query 62 VTGRRLKELMESNLRGEVT 80
++ KEL+ +LRGE+T
Sbjct 343 LSLDVRKELLTKDLRGEMT 361
> cel:F41C6.1 unc-6; UNCoordinated family member (unc-6); K06843
netrin 1
Length=612
Score = 34.7 bits (78), Expect = 0.074, Method: Composition-based stats.
Identities = 21/64 (32%), Positives = 30/64 (46%), Gaps = 3/64 (4%)
Query 8 AGVAVVAATQCHKGTANLLIYENGIWISSLGVICAKDMTPEAVVAKLYYLMGKGVTGRRL 67
A +V + +GT N+ E +WIS GVIC P+ V + Y L+GK +
Sbjct 514 AKYKIVVESVFKRGTENMQRGETSLWISPQGVICK---CPKLRVGRRYLLLGKNDSDHER 570
Query 68 KELM 71
LM
Sbjct 571 DGLM 574
Lambda K H
0.319 0.133 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2044675024
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40