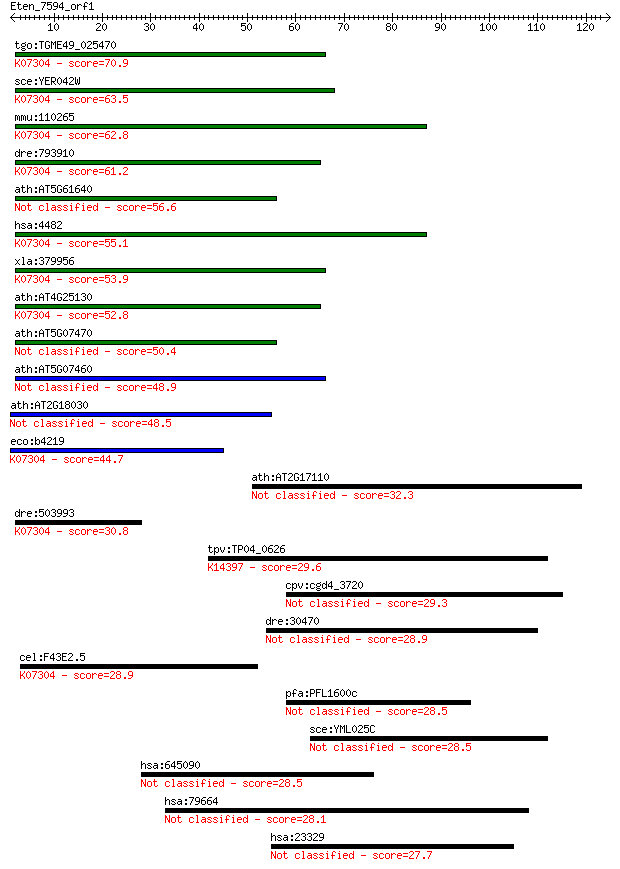
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7594_orf1
Length=124
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_025470 peptide methionine sulfoxide reductase, puta... 70.9 9e-13
sce:YER042W MXR1; Methionine-S-sulfoxide reductase, involved i... 63.5 1e-10
mmu:110265 Msra, 2310045J23Rik, 6530413P12Rik, MGC101976, MSR-... 62.8 3e-10
dre:793910 si:dkey-221h15.1 (EC:1.8.4.11); K07304 peptide-meth... 61.2 8e-10
ath:AT5G61640 PMSR1; PMSR1 (PEPTIDEMETHIONINE SULFOXIDE REDUCT... 56.6 2e-08
hsa:4482 MSRA, PMSR; methionine sulfoxide reductase A (EC:1.8.... 55.1 6e-08
xla:379956 msra.1, MGC64457, msra; methionine sulfoxide reduct... 53.9 1e-07
ath:AT4G25130 peptide methionine sulfoxide reductase, putative... 52.8 2e-07
ath:AT5G07470 PMSR3; PMSR3 (PEPTIDEMETHIONINE SULFOXIDE REDUCT... 50.4 1e-06
ath:AT5G07460 PMSR2; PMSR2 (PEPTIDEMETHIONINE SULFOXIDE REDUCT... 48.9 4e-06
ath:AT2G18030 peptide methionine sulfoxide reductase family pr... 48.5 5e-06
eco:b4219 msrA, ECK4215, JW4178, pms, pmsR; methionine sulfoxi... 44.7 8e-05
ath:AT2G17110 hypothetical protein 32.3 0.41
dre:503993 im:7149628; K07304 peptide-methionine (S)-S-oxide r... 30.8 1.2
tpv:TP04_0626 mRNA cleavage factor protein; K14397 cleavage an... 29.6 2.7
cpv:cgd4_3720 hypothetical protein 29.3 2.9
dre:30470 epb4.1l4, EPB41L4, nbl4, wu:fc29f06; erythrocyte pro... 28.9 4.1
cel:F43E2.5 hypothetical protein; K07304 peptide-methionine (S... 28.9 4.5
pfa:PFL1600c conserved Plasmodium protein 28.5 5.0
sce:YML025C YML6; Mitochondrial ribosomal protein of the large... 28.5 5.1
hsa:645090 CXorf30; chromosome X open reading frame 30 28.5
hsa:79664 NARG2, BRCC1; NMDA receptor regulated 2 28.1
hsa:23329 TBC1D30, KIAA0984; TBC1 domain family, member 30 27.7
> tgo:TGME49_025470 peptide methionine sulfoxide reductase, putative
(EC:1.8.4.11); K07304 peptide-methionine (S)-S-oxide
reductase [EC:1.8.4.11]
Length=269
Score = 70.9 bits (172), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 32/64 (50%), Positives = 42/64 (65%), Gaps = 0/64 (0%)
Query 2 LKISFFPQKVHLRELLTFFWTIHDPTTPDRQGPDVGPQYRSAVCVSDPTQLPLVLQIKNE 61
++I FF KV L +LL FFW IHDPTT +RQG DVG QY SA+ V ++++E
Sbjct 160 VEIQFFEDKVTLSDLLRFFWRIHDPTTLNRQGNDVGQQYGSAIFVYSAEHRQTAEELRDE 219
Query 62 MQKK 65
MQK+
Sbjct 220 MQKQ 223
> sce:YER042W MXR1; Methionine-S-sulfoxide reductase, involved
in the response to oxidative stress; protects iron-sulfur clusters
from oxidative inactivation along with MXR2; involved
in the regulation of lifespan (EC:1.8.4.11); K07304 peptide-methionine
(S)-S-oxide reductase [EC:1.8.4.11]
Length=184
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 41/66 (62%), Gaps = 0/66 (0%)
Query 2 LKISFFPQKVHLRELLTFFWTIHDPTTPDRQGPDVGPQYRSAVCVSDPTQLPLVLQIKNE 61
L++S+ P+ + LREL FF+ IHDPTT + QGPD G QYRS + L + +IK E
Sbjct 78 LQVSYNPKVITLRELTDFFFRIHDPTTSNSQGPDKGTQYRSGLFAHSDADLKELAKIKEE 137
Query 62 MQKKNG 67
Q K G
Sbjct 138 WQPKWG 143
> mmu:110265 Msra, 2310045J23Rik, 6530413P12Rik, MGC101976, MSR-A;
methionine sulfoxide reductase A (EC:1.8.4.11); K07304
peptide-methionine (S)-S-oxide reductase [EC:1.8.4.11]
Length=233
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 34/85 (40%), Positives = 45/85 (52%), Gaps = 4/85 (4%)
Query 2 LKISFFPQKVHLRELLTFFWTIHDPTTPDRQGPDVGPQYRSAVCVSDPTQLPLVLQIKNE 61
+++ + P+ + ELL FW HDPT RQG D G QYRSAV + Q+ L+ K E
Sbjct 117 VRVVYRPEHISFEELLKVFWENHDPTQGMRQGNDFGTQYRSAVYPTSAVQMEAALRSKEE 176
Query 62 MQKKNGISKSKLKSFPIPTNATSGQ 86
QK +SK PI T+ GQ
Sbjct 177 YQKV--LSKHNFG--PITTDIREGQ 197
> dre:793910 si:dkey-221h15.1 (EC:1.8.4.11); K07304 peptide-methionine
(S)-S-oxide reductase [EC:1.8.4.11]
Length=235
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 28/63 (44%), Positives = 37/63 (58%), Gaps = 0/63 (0%)
Query 2 LKISFFPQKVHLRELLTFFWTIHDPTTPDRQGPDVGPQYRSAVCVSDPTQLPLVLQIKNE 61
+++ F PQK+ ELL FW H+PT RQG DVG YRS++ + QL LQ + E
Sbjct 117 VRVVFEPQKIKFSELLKVFWESHNPTQGMRQGNDVGTTYRSSIYTNTQEQLEQALQSREE 176
Query 62 MQK 64
QK
Sbjct 177 YQK 179
> ath:AT5G61640 PMSR1; PMSR1 (PEPTIDEMETHIONINE SULFOXIDE REDUCTASE
1); oxidoreductase, acting on sulfur group of donors,
disulfide as acceptor / peptide-methionine-(S)-S-oxide reductase
Length=202
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 23/54 (42%), Positives = 34/54 (62%), Gaps = 0/54 (0%)
Query 2 LKISFFPQKVHLRELLTFFWTIHDPTTPDRQGPDVGPQYRSAVCVSDPTQLPLV 55
+++ + P++ + LL FW+ HDPTT +RQG DVG QYRS + +P Q L
Sbjct 89 VRVQYDPKECSYQSLLDLFWSKHDPTTLNRQGNDVGTQYRSGIYFYNPEQEKLA 142
> hsa:4482 MSRA, PMSR; methionine sulfoxide reductase A (EC:1.8.4.11);
K07304 peptide-methionine (S)-S-oxide reductase [EC:1.8.4.11]
Length=195
Score = 55.1 bits (131), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 43/85 (50%), Gaps = 4/85 (4%)
Query 2 LKISFFPQKVHLRELLTFFWTIHDPTTPDRQGPDVGPQYRSAVCVSDPTQLPLVLQIKNE 61
+++ + P+ + ELL FW HDPT RQG D G QYRSA+ + Q+ L K
Sbjct 79 VRVVYQPEHMSFEELLKVFWENHDPTQGMRQGNDHGTQYRSAIYPTSAKQMEAALSSKEN 138
Query 62 MQKKNGISKSKLKSFPIPTNATSGQ 86
QK +S+ PI T+ GQ
Sbjct 139 YQKV--LSEHGFG--PITTDIREGQ 159
> xla:379956 msra.1, MGC64457, msra; methionine sulfoxide reductase
A, gene 1 (EC:1.8.4.11); K07304 peptide-methionine (S)-S-oxide
reductase [EC:1.8.4.11]
Length=211
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 36/64 (56%), Gaps = 0/64 (0%)
Query 2 LKISFFPQKVHLRELLTFFWTIHDPTTPDRQGPDVGPQYRSAVCVSDPTQLPLVLQIKNE 61
+++ + P+ ++ +L+ FW HDPT RQG DVG YRSA+ Q L+ + +
Sbjct 96 VRVVYEPESINFEKLIKVFWENHDPTQGMRQGNDVGTMYRSAIYTYTKEQTEAALKSRED 155
Query 62 MQKK 65
QK+
Sbjct 156 YQKE 159
> ath:AT4G25130 peptide methionine sulfoxide reductase, putative;
K07304 peptide-methionine (S)-S-oxide reductase [EC:1.8.4.11]
Length=258
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 34/63 (53%), Gaps = 0/63 (0%)
Query 2 LKISFFPQKVHLRELLTFFWTIHDPTTPDRQGPDVGPQYRSAVCVSDPTQLPLVLQIKNE 61
+++ + P++ LL FW HDPTT +RQG DVG QYRS + Q + + +
Sbjct 145 VRVQYDPKECSFESLLDVFWNRHDPTTLNRQGGDVGTQYRSGIYYYTDEQERIAREAVEK 204
Query 62 MQK 64
QK
Sbjct 205 QQK 207
> ath:AT5G07470 PMSR3; PMSR3 (PEPTIDEMETHIONINE SULFOXIDE REDUCTASE
3); oxidoreductase, acting on sulfur group of donors,
disulfide as acceptor / peptide-methionine-(S)-S-oxide reductase
Length=202
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 22/54 (40%), Positives = 29/54 (53%), Gaps = 0/54 (0%)
Query 2 LKISFFPQKVHLRELLTFFWTIHDPTTPDRQGPDVGPQYRSAVCVSDPTQLPLV 55
+++ + LL FW+ HDPTT +RQG DVG QYRS + P Q L
Sbjct 89 VRVQYDLNDCTYESLLDLFWSRHDPTTLNRQGNDVGTQYRSGIYFYTPEQEKLA 142
> ath:AT5G07460 PMSR2; PMSR2 (PEPTIDEMETHIONINE SULFOXIDE REDUCTASE
2); oxidoreductase, acting on sulfur group of donors,
disulfide as acceptor / peptide-methionine-(S)-S-oxide reductase
Length=218
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 36/64 (56%), Gaps = 0/64 (0%)
Query 2 LKISFFPQKVHLRELLTFFWTIHDPTTPDRQGPDVGPQYRSAVCVSDPTQLPLVLQIKNE 61
+++ + P++ LL FW+ H+PTT +RQG +G QYRS + P Q L + +
Sbjct 105 VRVQYDPKECTYETLLDLFWSRHNPTTLNRQGELLGAQYRSGIYFYTPEQEKLARESLEK 164
Query 62 MQKK 65
QKK
Sbjct 165 EQKK 168
> ath:AT2G18030 peptide methionine sulfoxide reductase family
protein
Length=254
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 32/54 (59%), Gaps = 0/54 (0%)
Query 1 SLKISFFPQKVHLRELLTFFWTIHDPTTPDRQGPDVGPQYRSAVCVSDPTQLPL 54
S+++ + P+ + R+LL FW+ HD QGPDVG QYRS + + +L L
Sbjct 101 SVQVEYDPRIIGYRQLLDVFWSSHDSRQVFGQGPDVGNQYRSCIFTNSTEELRL 154
> eco:b4219 msrA, ECK4215, JW4178, pms, pmsR; methionine sulfoxide
reductase A (EC:1.8.4.11); K07304 peptide-methionine (S)-S-oxide
reductase [EC:1.8.4.11]
Length=212
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 19/44 (43%), Positives = 26/44 (59%), Gaps = 0/44 (0%)
Query 1 SLKISFFPQKVHLRELLTFFWTIHDPTTPDRQGPDVGPQYRSAV 44
+++I + P + +LL FW HDP RQG D G QYRSA+
Sbjct 96 AVRIVYDPSVISYEQLLQVFWENHDPAQGMRQGNDHGTQYRSAI 139
> ath:AT2G17110 hypothetical protein
Length=733
Score = 32.3 bits (72), Expect = 0.41, Method: Composition-based stats.
Identities = 20/69 (28%), Positives = 38/69 (55%), Gaps = 1/69 (1%)
Query 51 QLPLVLQI-KNEMQKKNGISKSKLKSFPIPTNATSGQPKTTTKSTSSRTLAATAPTGSTS 109
++ ++L++ K+ +KN SK + P P+ +S Q T+ K+ + + + TAPT +
Sbjct 331 EIAVMLEVGKHPYGRKNVSSKKLYEGTPSPSVVSSAQSSTSKKAKAEASSSVTAPTYADI 390
Query 110 ELQLRLLLR 118
E +L L R
Sbjct 391 EAELALKSR 399
> dre:503993 im:7149628; K07304 peptide-methionine (S)-S-oxide
reductase [EC:1.8.4.11]
Length=160
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 2 LKISFFPQKVHLRELLTFFWTIHDPT 27
+++ F P+ V L +LL FW HDPT
Sbjct 134 VRVVFSPKDVSLEKLLKVFWESHDPT 159
> tpv:TP04_0626 mRNA cleavage factor protein; K14397 cleavage
and polyadenylation specificity factor subunit 5
Length=226
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 29/73 (39%), Gaps = 3/73 (4%)
Query 42 SAVCVSDPTQLPLVLQIKNEMQKKNGISKSKLKSFPIPTNATSGQPK---TTTKSTSSRT 98
V +S P VL +K ++ K G+ K KSF P S + T+TK
Sbjct 70 CGVILSHRKGFPFVLLLKRDLDKSVGLLGGKCKSFENPKEVLSSKLARFITSTKHKHQLN 129
Query 99 LAATAPTGSTSEL 111
+ T T EL
Sbjct 130 IKETIETIQVGEL 142
> cpv:cgd4_3720 hypothetical protein
Length=1317
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 26/57 (45%), Gaps = 0/57 (0%)
Query 58 IKNEMQKKNGISKSKLKSFPIPTNATSGQPKTTTKSTSSRTLAATAPTGSTSELQLR 114
I+ E + ++ +K K K I +N P T T + + + P G S LQLR
Sbjct 1195 IEFEFRVEDNFTKLKFKINAINSNNGENTPSTITSDDAENSPSILPPLGDISYLQLR 1251
> dre:30470 epb4.1l4, EPB41L4, nbl4, wu:fc29f06; erythrocyte protein
band 4.1-like 4
Length=619
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 27/57 (47%), Gaps = 1/57 (1%)
Query 54 LVLQIKNEMQKKNGISKSKLKSFPIPTNATSGQPKTTTKSTSSRTLAAT-APTGSTS 109
L I+ E+ +G+S+ +LK P T G P S S R+ +T APT T
Sbjct 544 LWKHIQKELVDPSGLSEEQLKEIPYKKVETQGDPIKIRHSHSPRSFGSTGAPTARTE 600
> cel:F43E2.5 hypothetical protein; K07304 peptide-methionine
(S)-S-oxide reductase [EC:1.8.4.11]
Length=207
Score = 28.9 bits (63), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 14/49 (28%), Positives = 25/49 (51%), Gaps = 6/49 (12%)
Query 3 KISFFPQKVHLRELLTFFWTIHDPTTPDRQGPDVGPQYRSAVCVSDPTQ 51
+I+F P+ + +L FFW H+P ++ QY+SA+ + Q
Sbjct 54 EITFDPKVIEYSKLTNFFWKHHNPAERRKK------QYQSAILYVNDDQ 96
> pfa:PFL1600c conserved Plasmodium protein
Length=3193
Score = 28.5 bits (62), Expect = 5.0, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 20/38 (52%), Gaps = 2/38 (5%)
Query 58 IKNEMQKKNGISKSKLKSFPIPTNATSGQPKTTTKSTS 95
+KNE + K S SFPI TN + K T K+T+
Sbjct 699 LKNETENKT--RNSTYNSFPINTNTEQNKSKITRKNTN 734
> sce:YML025C YML6; Mitochondrial ribosomal protein of the large
subunit, has similarity to E. coli L4 ribosomal protein and
human mitoribosomal MRP-L4 protein; essential for viability,
unlike most other mitoribosomal proteins
Length=286
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 23/49 (46%), Gaps = 0/49 (0%)
Query 63 QKKNGISKSKLKSFPIPTNATSGQPKTTTKSTSSRTLAATAPTGSTSEL 111
+ +NG S+ KL A G + T+ R LA TAP T+EL
Sbjct 95 RSENGFSRRKLMPQKGSGRARVGDANSPTRHNGGRALARTAPNDYTTEL 143
> hsa:645090 CXorf30; chromosome X open reading frame 30
Length=633
Score = 28.5 bits (62), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 26/48 (54%), Gaps = 0/48 (0%)
Query 28 TPDRQGPDVGPQYRSAVCVSDPTQLPLVLQIKNEMQKKNGISKSKLKS 75
TPD V P+ S DP ++P + + + E+Q ++ KSKL+S
Sbjct 374 TPDLTEVLVIPKRNSHNFCEDPNEIPKIHEFEYEIQFESEAMKSKLES 421
> hsa:79664 NARG2, BRCC1; NMDA receptor regulated 2
Length=845
Score = 28.1 bits (61), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 34/77 (44%), Gaps = 7/77 (9%)
Query 33 GPDVGPQYRSAVCVSDPTQLPLVL--QIKNEMQKKNGISKSKLKSFPIPTNATSGQPKTT 90
G + + +C SD + L++ + KN K + S L S P N++SGQ
Sbjct 420 GSEAAKTEDTVLCSSDTDEECLIIDTECKNNSDGKTAVVGSNLSSRPASPNSSSGQA--- 476
Query 91 TKSTSSRTLAATAPTGS 107
S ++T A +P S
Sbjct 477 --SVGNQTNTACSPEES 491
> hsa:23329 TBC1D30, KIAA0984; TBC1 domain family, member 30
Length=761
Score = 27.7 bits (60), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 13/50 (26%), Positives = 29/50 (58%), Gaps = 2/50 (4%)
Query 55 VLQIKNEMQKKNGISKSKLKSFPIPTNATSGQPKTTTKSTSSRTLAATAP 104
++ +NE K NG+ ++ +FP AT+G+ ++ + ++ RT+ +P
Sbjct 591 LIDEQNEASKTNGLGAAE--AFPSGCTATAGREGSSPEGSTRRTIEGQSP 638
Lambda K H
0.314 0.130 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2069971060
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40