bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7615_orf3
Length=75
Score E
Sequences producing significant alignments: (Bits) Value
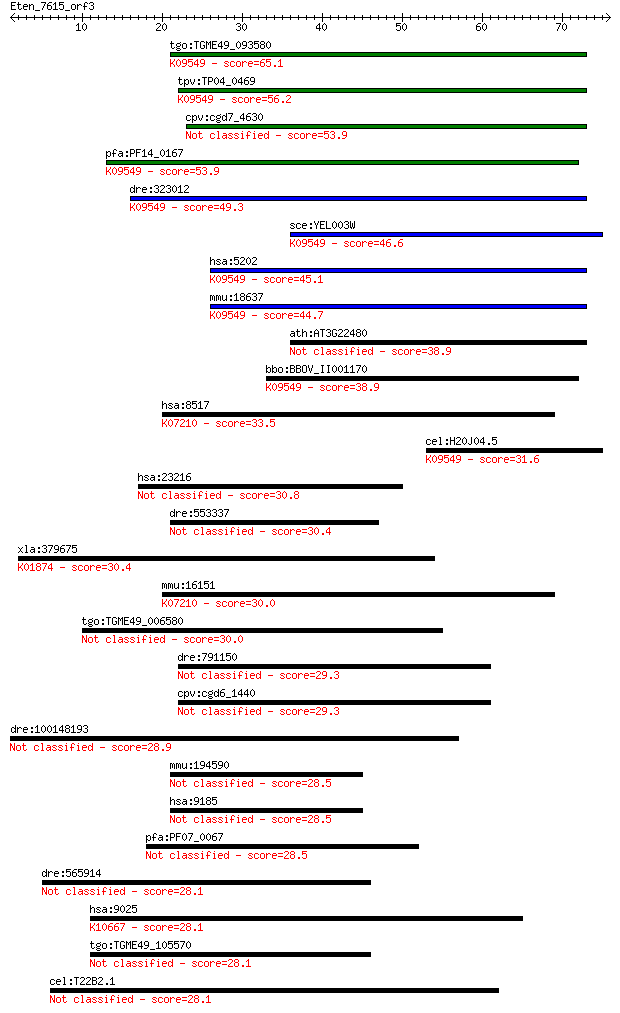
tgo:TGME49_093580 hypothetical protein ; K09549 prefoldin subu... 65.1 5e-11
tpv:TP04_0469 hypothetical protein; K09549 prefoldin subunit 2 56.2
cpv:cgd7_4630 hypothetical protein 53.9 1e-07
pfa:PF14_0167 prefoldin subunit 2, putative; K09549 prefoldin ... 53.9 1e-07
dre:323012 pfdn2, im:6895552, wu:fb79a01, zgc:136473; prefoldi... 49.3 3e-06
sce:YEL003W GIM4, PFD2; Gim4p; K09549 prefoldin subunit 2 46.6
hsa:5202 PFDN2, PFD2; prefoldin subunit 2; K09549 prefoldin su... 45.1 6e-05
mmu:18637 Pfdn2, ESTM27, W48336; prefoldin 2; K09549 prefoldin... 44.7 8e-05
ath:AT3G22480 PDF2; prefoldin-related KE2 family protein 38.9 0.003
bbo:BBOV_II001170 18.m06087; KE2 family protein; K09549 prefol... 38.9 0.004
hsa:8517 IKBKG, AMCBX1, FIP-3, FIP3, Fip3p, IKK-gamma, IP, IP1... 33.5 0.16
cel:H20J04.5 pfd-2; PreFolDin (molecular chaperone) family mem... 31.6 0.68
hsa:23216 TBC1D1, KIAA1108, TBC, TBC1; TBC1 (tre-2/USP6, BUB2,... 30.8 1.1
dre:553337 lamb2l, fc83a09, si:dkey-32e6.1, wu:fc83a09; lamini... 30.4 1.3
xla:379675 mars, MGC69150; methionyl-tRNA synthetase, cytoplas... 30.4 1.5
mmu:16151 Ikbkg, 1110037D23Rik, AI848108, AI851264, AW124339, ... 30.0 1.7
tgo:TGME49_006580 hypothetical protein 30.0 2.0
dre:791150 gripap1, zgc:158787; GRIP1 associated protein 1 29.3
cpv:cgd6_1440 hypothetical protein 29.3 2.9
dre:100148193 dnah6; dynein, axonemal, heavy chain 6 28.9
mmu:194590 Reps2, POB1; RALBP1 associated Eps domain containin... 28.5 4.8
hsa:9185 REPS2, POB1; RALBP1 associated Eps domain containing 2 28.5
pfa:PF07_0067 conserved Plasmodium protein, unknown function 28.5 5.3
dre:565914 plekhh1, im:6897489, im:7143671, max1; pleckstrin h... 28.1 6.1
hsa:9025 RNF8, FLJ12013, KIAA0646; ring finger protein 8 (EC:6... 28.1 6.4
tgo:TGME49_105570 hypothetical protein 28.1 6.6
cel:T22B2.1 hypothetical protein 28.1 7.2
> tgo:TGME49_093580 hypothetical protein ; K09549 prefoldin subunit
2
Length=170
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 27/52 (51%), Positives = 42/52 (80%), Gaps = 0/52 (0%)
Query 21 DLKQRLQRLEKERIALNNTIEELRQDSSDHQLVLDAFKDLEPSRRCYRLVGG 72
+L+Q L R+E+E++ L + I++L+QD+S+H+LVLDAF + RRCYR+VGG
Sbjct 38 ELQQALHRVEREKVILVSKIQDLQQDTSEHRLVLDAFAKVNADRRCYRMVGG 89
> tpv:TP04_0469 hypothetical protein; K09549 prefoldin subunit
2
Length=114
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 25/51 (49%), Positives = 36/51 (70%), Gaps = 0/51 (0%)
Query 22 LKQRLQRLEKERIALNNTIEELRQDSSDHQLVLDAFKDLEPSRRCYRLVGG 72
+K L+RLEKER+ L +E++ QDSS+H+LVL L R+CYR++GG
Sbjct 7 MKATLKRLEKERVTLLGQMEDVSQDSSEHRLVLKNLSKLPSDRKCYRIIGG 57
> cpv:cgd7_4630 hypothetical protein
Length=132
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 21/50 (42%), Positives = 39/50 (78%), Gaps = 0/50 (0%)
Query 23 KQRLQRLEKERIALNNTIEELRQDSSDHQLVLDAFKDLEPSRRCYRLVGG 72
++++ +L+++ L++ I EL QDS +H LVL+AF+ ++P RRC+R++GG
Sbjct 14 REKISQLDRKLKVLSSKISELSQDSKEHGLVLNAFEKVDPERRCFRVIGG 63
> pfa:PF14_0167 prefoldin subunit 2, putative; K09549 prefoldin
subunit 2
Length=147
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 25/59 (42%), Positives = 41/59 (69%), Gaps = 0/59 (0%)
Query 13 APRPSGEEDLKQRLQRLEKERIALNNTIEELRQDSSDHQLVLDAFKDLEPSRRCYRLVG 71
+P S ++ K +++EK+R+ L + IEEL QD +H+LVL+A +++ RRCYR+VG
Sbjct 9 SPNISEPKESKLTYEQIEKDRVQLVSKIEELYQDVVEHKLVLEALENVPSDRRCYRMVG 67
> dre:323012 pfdn2, im:6895552, wu:fb79a01, zgc:136473; prefoldin
subunit 2; K09549 prefoldin subunit 2
Length=156
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 21/57 (36%), Positives = 38/57 (66%), Gaps = 1/57 (1%)
Query 16 PSGEEDLKQRLQRLEKERIALNNTIEELRQDSSDHQLVLDAFKDLEPSRRCYRLVGG 72
PS E+ + QR+ +E+ ++ + E + ++H LV+D K+++PSR+C+RLVGG
Sbjct 20 PSAEQ-VVATFQRMRQEQRSMASKAAEFEMEINEHSLVIDTLKEVDPSRKCFRLVGG 75
> sce:YEL003W GIM4, PFD2; Gim4p; K09549 prefoldin subunit 2
Length=111
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 17/39 (43%), Positives = 24/39 (61%), Gaps = 0/39 (0%)
Query 36 LNNTIEELRQDSSDHQLVLDAFKDLEPSRRCYRLVGGGF 74
L I EL D +H +V+ KD EP+R+CYR++GG
Sbjct 22 LQTKIIELGHDKDEHTIVIKTLKDAEPTRKCYRMIGGAL 60
> hsa:5202 PFDN2, PFD2; prefoldin subunit 2; K09549 prefoldin
subunit 2
Length=154
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 31/47 (65%), Gaps = 0/47 (0%)
Query 26 LQRLEKERIALNNTIEELRQDSSDHQLVLDAFKDLEPSRRCYRLVGG 72
RL +E+ L + EL + ++H LV+D K+++ +R+CYR+VGG
Sbjct 30 FNRLRQEQRGLASKAAELEMELNEHSLVIDTLKEVDETRKCYRMVGG 76
> mmu:18637 Pfdn2, ESTM27, W48336; prefoldin 2; K09549 prefoldin
subunit 2
Length=154
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 31/47 (65%), Gaps = 0/47 (0%)
Query 26 LQRLEKERIALNNTIEELRQDSSDHQLVLDAFKDLEPSRRCYRLVGG 72
RL +E+ L + EL + ++H LV+D K+++ +R+CYR+VGG
Sbjct 30 FNRLRQEQRGLASKAAELEMELNEHSLVIDTLKEVDETRKCYRMVGG 76
> ath:AT3G22480 PDF2; prefoldin-related KE2 family protein
Length=148
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 26/37 (70%), Gaps = 0/37 (0%)
Query 36 LNNTIEELRQDSSDHQLVLDAFKDLEPSRRCYRLVGG 72
+ + I +L S+H LV++A + L+ SR+C+R++GG
Sbjct 33 IYSNITDLEMQVSEHSLVINAIQPLDQSRKCFRMIGG 69
> bbo:BBOV_II001170 18.m06087; KE2 family protein; K09549 prefoldin
subunit 2
Length=109
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 17/39 (43%), Positives = 26/39 (66%), Gaps = 0/39 (0%)
Query 33 RIALNNTIEELRQDSSDHQLVLDAFKDLEPSRRCYRLVG 71
R+ L +++L Q+S++H+LVL L RRCYR+VG
Sbjct 18 RLRLLGQLDDLSQESAEHKLVLKTLSTLPDDRRCYRIVG 56
> hsa:8517 IKBKG, AMCBX1, FIP-3, FIP3, Fip3p, IKK-gamma, IP, IP1,
IP2, IPD2, NEMO; inhibitor of kappa light polypeptide gene
enhancer in B-cells, kinase gamma; K07210 inhibitor of nuclear
factor kappa-B kinase subunit gamma
Length=487
Score = 33.5 bits (75), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 31/49 (63%), Gaps = 0/49 (0%)
Query 20 EDLKQRLQRLEKERIALNNTIEELRQDSSDHQLVLDAFKDLEPSRRCYR 68
EDLKQ+LQ+ E+ +A I++L++++ H++V++ L+ Y+
Sbjct 329 EDLKQQLQQAEEALVAKQEVIDKLKEEAEQHKIVMETVPVLKAQADIYK 377
> cel:H20J04.5 pfd-2; PreFolDin (molecular chaperone) family member
(pfd-2); K09549 prefoldin subunit 2
Length=141
Score = 31.6 bits (70), Expect = 0.68, Method: Compositional matrix adjust.
Identities = 10/22 (45%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 53 VLDAFKDLEPSRRCYRLVGGGF 74
VL+ KDLEP ++C+RL+
Sbjct 51 VLEVIKDLEPDQKCFRLISDTL 72
> hsa:23216 TBC1D1, KIAA1108, TBC, TBC1; TBC1 (tre-2/USP6, BUB2,
cdc16) domain family, member 1
Length=1168
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 17/33 (51%), Positives = 22/33 (66%), Gaps = 0/33 (0%)
Query 17 SGEEDLKQRLQRLEKERIALNNTIEELRQDSSD 49
S E LKQ + LE ER AL T+EELR+ S++
Sbjct 1120 SSESKLKQAMLTLELERSALLQTVEELRRRSAE 1152
> dre:553337 lamb2l, fc83a09, si:dkey-32e6.1, wu:fc83a09; laminin,
beta 2-like
Length=1838
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 15/26 (57%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 21 DLKQRLQRLEKERIALNNTIEELRQD 46
DLKQ L LEKE LNNT+ +LR++
Sbjct 1324 DLKQNLTALEKELKDLNNTLHQLRRE 1349
> xla:379675 mars, MGC69150; methionyl-tRNA synthetase, cytoplasmic
(EC:6.1.1.10); K01874 methionyl-tRNA synthetase [EC:6.1.1.10]
Length=905
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 30/52 (57%), Gaps = 4/52 (7%)
Query 2 LNPRQMAQPQGAPRPSGEEDLKQRLQRLEKERIALNNTIEELRQDSSDHQLV 53
++P Q A Q AP+ SG E +K+ +Q LEK+ N + EL+ ++ ++
Sbjct 825 VSPSQEAPEQQAPKASGPERVKELMQELEKQ----GNHVRELKGKKAEKSVI 872
> mmu:16151 Ikbkg, 1110037D23Rik, AI848108, AI851264, AW124339,
IKK[g], NEMO; inhibitor of kappaB kinase gamma; K07210 inhibitor
of nuclear factor kappa-B kinase subunit gamma
Length=430
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 14/49 (28%), Positives = 31/49 (63%), Gaps = 0/49 (0%)
Query 20 EDLKQRLQRLEKERIALNNTIEELRQDSSDHQLVLDAFKDLEPSRRCYR 68
EDL+Q+LQ+ E+ +A I++L++++ H++V++ L+ Y+
Sbjct 272 EDLRQQLQQAEEALVAKQELIDKLKEEAEQHKIVMETVPVLKAQADIYK 320
> tgo:TGME49_006580 hypothetical protein
Length=4740
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 27/50 (54%), Gaps = 5/50 (10%)
Query 10 PQGAPRPSG-----EEDLKQRLQRLEKERIALNNTIEELRQDSSDHQLVL 54
P GAPRPSG +E L L RL+K + T+E LR+ + + V+
Sbjct 3822 PSGAPRPSGSKPQTDEALHAELTRLQKLHGEMVLTLERLRESYLNCKAVI 3871
> dre:791150 gripap1, zgc:158787; GRIP1 associated protein 1
Length=867
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 20/47 (42%), Positives = 30/47 (63%), Gaps = 8/47 (17%)
Query 22 LKQRLQRLEKERIALNNTIEELRQ---DSSDHQLVLD-----AFKDL 60
++Q++ +EKER LNNTI +L+Q D+ D Q +L+ A KDL
Sbjct 594 VQQQVTEMEKEREDLNNTIGKLKQEIKDTVDGQRILEKKGSSALKDL 640
> cpv:cgd6_1440 hypothetical protein
Length=711
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 24/39 (61%), Gaps = 0/39 (0%)
Query 22 LKQRLQRLEKERIALNNTIEELRQDSSDHQLVLDAFKDL 60
+ RL +LE E+ +LNN I EL ++++L +D + L
Sbjct 385 INSRLSKLEIEKKSLNNYITELETSKAEYKLKIDELEHL 423
> dre:100148193 dnah6; dynein, axonemal, heavy chain 6
Length=4163
Score = 28.9 bits (63), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 34/59 (57%), Gaps = 3/59 (5%)
Query 1 TLNPRQMAQPQGAPRPSGEED--LKQRLQRLEKERIALNNTIEELRQDSSDHQLV-LDA 56
TL P A + GE D + + Q L+++ LNNT++E++ ++ +HQL+ +DA
Sbjct 787 TLPPCITAVRNTIDKAVGERDSYVDKFCQHLQQDIGQLNNTVQEVKAEAENHQLLDIDA 845
> mmu:194590 Reps2, POB1; RALBP1 associated Eps domain containing
protein 2
Length=648
Score = 28.5 bits (62), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 12/24 (50%), Positives = 20/24 (83%), Gaps = 0/24 (0%)
Query 21 DLKQRLQRLEKERIALNNTIEELR 44
+L+Q+L+ + +ERIAL N +E+LR
Sbjct 620 ELQQQLKEVHQERIALENQLEQLR 643
> hsa:9185 REPS2, POB1; RALBP1 associated Eps domain containing
2
Length=659
Score = 28.5 bits (62), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 12/24 (50%), Positives = 20/24 (83%), Gaps = 0/24 (0%)
Query 21 DLKQRLQRLEKERIALNNTIEELR 44
+L+Q+L+ + +ERIAL N +E+LR
Sbjct 631 ELQQQLKEVHQERIALENQLEQLR 654
> pfa:PF07_0067 conserved Plasmodium protein, unknown function
Length=1069
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 15/35 (42%), Positives = 23/35 (65%), Gaps = 1/35 (2%)
Query 18 GEEDLKQRLQRLEKERIALNNTIEELRQDSSD-HQ 51
G+ D K+ + ++EKE+I N IEEL+ S+ HQ
Sbjct 407 GKYDNKELVVKIEKEKIDKNKKIEELKNISNQTHQ 441
> dre:565914 plekhh1, im:6897489, im:7143671, max1; pleckstrin
homology domain containing, family H (with MyTH4 domain) member
1
Length=1433
Score = 28.1 bits (61), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 13/41 (31%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 5 RQMAQPQGAPRPSGEEDLKQRLQRLEKERIALNNTIEELRQ 45
R P G+P+P ++ L Q L+R +R ++E+LR+
Sbjct 1213 RAAMSPVGSPQPKTQQTLLQVLERFYPKRYKQECSVEQLRE 1253
> hsa:9025 RNF8, FLJ12013, KIAA0646; ring finger protein 8 (EC:6.3.2.-);
K10667 E3 ubiquitin-protein ligase RNF8 [EC:6.3.2.19]
Length=485
Score = 28.1 bits (61), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 32/55 (58%), Gaps = 4/55 (7%)
Query 11 QGAPRPSGEEDLKQRLQRLEKERIALNNTIEELRQDSSDHQLVLDA-FKDLEPSR 64
QG GE+DLKQ+L + +E AL +EEL + D + ++ A K+LE ++
Sbjct 325 QGLEIAQGEKDLKQQLAQALQEHWAL---MEELNRSKKDFEAIIQAKNKELEQTK 376
> tgo:TGME49_105570 hypothetical protein
Length=336
Score = 28.1 bits (61), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 11 QGAPRPSGEEDLKQRLQRLEKERIALNNTIEELRQ 45
Q G + K +QRLEKE+ L IE+L+Q
Sbjct 129 QAESSSGGFSECKHEIQRLEKEKFLLCEKIEKLKQ 163
> cel:T22B2.1 hypothetical protein
Length=955
Score = 28.1 bits (61), Expect = 7.2, Method: Composition-based stats.
Identities = 17/56 (30%), Positives = 29/56 (51%), Gaps = 0/56 (0%)
Query 6 QMAQPQGAPRPSGEEDLKQRLQRLEKERIALNNTIEELRQDSSDHQLVLDAFKDLE 61
Q Q A + + LK++ Q+LE++R NNT L + H++++D K E
Sbjct 730 QSQQTVIAQQEKSIKSLKEKSQQLEEKRSITNNTQRLLEETEIIHKILVDTLKAQE 785
Lambda K H
0.316 0.136 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2002740660
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40