bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7664_orf1
Length=153
Score E
Sequences producing significant alignments: (Bits) Value
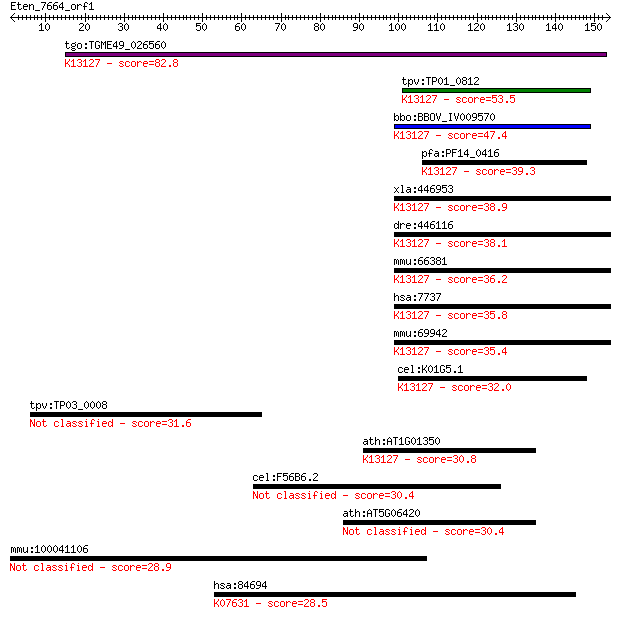
tgo:TGME49_026560 zinc finger (CCCH type) protein, putative (E... 82.8 4e-16
tpv:TP01_0812 hypothetical protein; K13127 RING finger protein... 53.5 3e-07
bbo:BBOV_IV009570 23.m05882; zinc finger protein; K13127 RING ... 47.4 2e-05
pfa:PF14_0416 zinc finger protein, putative; K13127 RING finge... 39.3 0.006
xla:446953 zinc finger protein 183; K13127 RING finger protein... 38.9 0.006
dre:446116 rnf113a, wu:fj91a07, zgc:110007, znf183; ring finge... 38.1 0.012
mmu:66381 Rnf113a2, 2310020H19Rik, AI426758, MGC144613, Rnf113... 36.2 0.043
hsa:7737 RNF113A, Cwc24, RNF113, ZNF183; ring finger protein 1... 35.8 0.048
mmu:69942 Rnf113a1, 2810428C21Rik, Rnf113a; ring finger protei... 35.4 0.068
cel:K01G5.1 rnf-113; RiNg Finger protein family member (rnf-11... 32.0 0.79
tpv:TP03_0008 hypothetical protein 31.6 1.0
ath:AT1G01350 nucleic acid binding / protein binding / zinc io... 30.8 1.8
cel:F56B6.2 rgs-7; Regulator of G protein Signaling family mem... 30.4 2.4
ath:AT5G06420 zinc finger (CCCH-type/C3HC4-type RING finger) f... 30.4 2.5
mmu:100041106 Gm3141; predicted gene 3141 28.9
hsa:84694 GJA10, CX62, RP11-63K6.6; gap junction protein, alph... 28.5 8.4
> tgo:TGME49_026560 zinc finger (CCCH type) protein, putative
(EC:4.1.2.13); K13127 RING finger protein 113A
Length=411
Score = 82.8 bits (203), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 58/153 (37%), Positives = 77/153 (50%), Gaps = 18/153 (11%)
Query 15 MFAKRAVPKGVRKRPNPNTPE------GPPQGTCG---------SDGSDEEHAKIVKPRN 59
MFAKRA K R+ P E G GT SD DE+ +VK +
Sbjct 1 MFAKRATKKQGRRGPLSEEAEAEATAGGSASGTQTTRAEKDERQSDKDDEDPVHVVKKQR 60
Query 60 PRKLLCKHFLVGKTKGEKGEQQKATDALAQQSNPDLGLDDDNRATATLEVDAGRGEDKRA 119
KHF+ ++ K ++ +SNP L +++DNRATA +VD R D RA
Sbjct 61 TASTG-KHFIQAVSERTKHTAEEMLHGF--KSNPKLTINNDNRATAIFDVDTDRKHDHRA 117
Query 120 LWERRLAISETLKKGLLEPGVYRGQGAHRLYAK 152
+ ER I E ++KG LE G+YRGQGAHR+Y K
Sbjct 118 ILERNAEIGEKIEKGELEAGIYRGQGAHRVYVK 150
> tpv:TP01_0812 hypothetical protein; K13127 RING finger protein
113A
Length=328
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 23/48 (47%), Positives = 32/48 (66%), Gaps = 0/48 (0%)
Query 101 NRATATLEVDAGRGEDKRALWERRLAISETLKKGLLEPGVYRGQGAHR 148
NRAT+T E+D R D R++ ER L I + G LEP +YRG+GA++
Sbjct 55 NRATSTYEIDTDRSMDHRSILERNLEIGNKILAGELEPNIYRGKGAYK 102
> bbo:BBOV_IV009570 23.m05882; zinc finger protein; K13127 RING
finger protein 113A
Length=319
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 22/50 (44%), Positives = 31/50 (62%), Gaps = 0/50 (0%)
Query 99 DDNRATATLEVDAGRGEDKRALWERRLAISETLKKGLLEPGVYRGQGAHR 148
+D RAT+T E+D D R++ ER I + + LE GVYRG+GA+R
Sbjct 46 NDQRATSTYEIDTEVSRDHRSILERNAEIGKQILNNELEDGVYRGRGAYR 95
> pfa:PF14_0416 zinc finger protein, putative; K13127 RING finger
protein 113A
Length=408
Score = 39.3 bits (90), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 25/42 (59%), Gaps = 0/42 (0%)
Query 106 TLEVDAGRGEDKRALWERRLAISETLKKGLLEPGVYRGQGAH 147
+ E+D D R++ ER + I E + KG L+ +YRG+ AH
Sbjct 137 SYEIDQDIKNDHRSIMERNIKIGEEILKGNLKDNIYRGKDAH 178
> xla:446953 zinc finger protein 183; K13127 RING finger protein
113A
Length=319
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 33/55 (60%), Gaps = 1/55 (1%)
Query 99 DDNRATATLEVDAGRGEDKRALWERRLAISETLKKGLLEPGVYRGQGAHRLYAKP 153
DD ATAT E+D + +D +A++ER + E + KG + +YRG ++ + KP
Sbjct 97 DDMGATATYELDTEKDKDAQAIFERSQKVQEEI-KGKEDDKIYRGIHNYQKFVKP 150
> dre:446116 rnf113a, wu:fj91a07, zgc:110007, znf183; ring finger
protein 113A; K13127 RING finger protein 113A
Length=321
Score = 38.1 bits (87), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 21/55 (38%), Positives = 30/55 (54%), Gaps = 1/55 (1%)
Query 99 DDNRATATLEVDAGRGEDKRALWERRLAISETLKKGLLEPGVYRGQGAHRLYAKP 153
DD ATA E+D R +D +A++ER I E L G + +YRG + + KP
Sbjct 100 DDMGATAVYELDTERDKDAQAIFERSQKIQEEL-TGKEDDKIYRGINNYHKFIKP 153
> mmu:66381 Rnf113a2, 2310020H19Rik, AI426758, MGC144613, Rnf113a;
ring finger protein 113A2; K13127 RING finger protein 113A
Length=337
Score = 36.2 bits (82), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 31/55 (56%), Gaps = 1/55 (1%)
Query 99 DDNRATATLEVDAGRGEDKRALWERRLAISETLKKGLLEPGVYRGQGAHRLYAKP 153
+D ATA E+D + D +A++ER I E L +G + +YRG ++ Y KP
Sbjct 112 EDMGATAVYELDTEKERDAQAIFERSQKIQEEL-RGKEDDKIYRGINNYQKYMKP 165
> hsa:7737 RNF113A, Cwc24, RNF113, ZNF183; ring finger protein
113A; K13127 RING finger protein 113A
Length=343
Score = 35.8 bits (81), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 31/55 (56%), Gaps = 1/55 (1%)
Query 99 DDNRATATLEVDAGRGEDKRALWERRLAISETLKKGLLEPGVYRGQGAHRLYAKP 153
+D ATA E+D + D +A++ER I E L +G + +YRG ++ Y KP
Sbjct 112 EDMGATAVYELDTEKERDAQAIFERSQKIQEEL-RGKEDDKIYRGINNYQKYMKP 165
> mmu:69942 Rnf113a1, 2810428C21Rik, Rnf113a; ring finger protein
113A1; K13127 RING finger protein 113A
Length=341
Score = 35.4 bits (80), Expect = 0.068, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 31/55 (56%), Gaps = 1/55 (1%)
Query 99 DDNRATATLEVDAGRGEDKRALWERRLAISETLKKGLLEPGVYRGQGAHRLYAKP 153
+D ATA E+D + D ++++ER I E L +G + +YRG ++ Y KP
Sbjct 110 EDMGATAVYELDTEKDRDAQSIFERSQKIQEEL-RGKEDDKIYRGINNYQKYVKP 163
> cel:K01G5.1 rnf-113; RiNg Finger protein family member (rnf-113);
K13127 RING finger protein 113A
Length=384
Score = 32.0 bits (71), Expect = 0.79, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 29/50 (58%), Gaps = 5/50 (10%)
Query 100 DNRATATLEVDAGRGEDKRALWERRLAISETLKKGLLEPG--VYRGQGAH 147
D ATATLEVD D +A +ER + + LK+G+ + G +Y+G +
Sbjct 93 DQGATATLEVDTDYSHDAQAQFER---VQQQLKEGVEKDGKILYKGSALY 139
> tpv:TP03_0008 hypothetical protein
Length=911
Score = 31.6 bits (70), Expect = 1.0, Method: Composition-based stats.
Identities = 21/60 (35%), Positives = 31/60 (51%), Gaps = 9/60 (15%)
Query 6 PAPGLPSPQMFAKRAVPKGVRKRPNPNTPEGPPQGTCGSDGSDEEHAKIV-KPRNPRKLL 64
P PG PSP PKG K P P TP+ P T D +++++ + +PR P ++L
Sbjct 198 PTPG-PSPLK------PKG-DKEPAPTTPKEPVPVTPKRDEDEDDYSPVTPRPREPVEVL 249
> ath:AT1G01350 nucleic acid binding / protein binding / zinc
ion binding; K13127 RING finger protein 113A
Length=343
Score = 30.8 bits (68), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 27/44 (61%), Gaps = 0/44 (0%)
Query 91 SNPDLGLDDDNRATATLEVDAGRGEDKRALWERRLAISETLKKG 134
S+ ++ + +D+ ATATLE + +D RA+ ER L ++ KG
Sbjct 102 SSKEIQVQNDSGATATLETETDFNQDARAIRERVLKKADEALKG 145
> cel:F56B6.2 rgs-7; Regulator of G protein Signaling family member
(rgs-7)
Length=819
Score = 30.4 bits (67), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 34/67 (50%), Gaps = 5/67 (7%)
Query 63 LLCKHFLVGKTKGEKGEQQKA----TDALAQQSNPDLGLDDDNRATATLEVDAGRGEDKR 118
L C+ F K G+K QKA ++ +A+ S ++ LD D RA V+AG D
Sbjct 713 LECEEFKKMK-DGKKSTTQKAIEIYSEFVAEHSPKEVNLDSDTRAATKAAVEAGCKPDTF 771
Query 119 ALWERRL 125
AL + R+
Sbjct 772 ALAQSRV 778
> ath:AT5G06420 zinc finger (CCCH-type/C3HC4-type RING finger)
family protein
Length=378
Score = 30.4 bits (67), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 27/49 (55%), Gaps = 0/49 (0%)
Query 86 ALAQQSNPDLGLDDDNRATATLEVDAGRGEDKRALWERRLAISETLKKG 134
S+ ++ + +D+ ATATLE + +D RA+ ER L ++ KG
Sbjct 132 VFHYDSSKEIQVQNDSGATATLETETDFNQDARAIRERVLKKADHALKG 180
> mmu:100041106 Gm3141; predicted gene 3141
Length=521
Score = 28.9 bits (63), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 28/107 (26%), Positives = 45/107 (42%), Gaps = 6/107 (5%)
Query 1 TLNPRPAPGLPSPQMFAKRAVPKGVRKRPNPNTPEGPPQGTCGSDG-SDEEHAKIVKPRN 59
T+ P P +PS P G R TP+GPP+ + S + AK V+ R+
Sbjct 417 TVTETPEPAMPSGVY-----RPPGARLTTTRKTPQGPPEIYSDTQFPSLQSTAKHVESRD 471
Query 60 PRKLLCKHFLVGKTKGEKGEQQKATDALAQQSNPDLGLDDDNRATAT 106
K + K F V + K E+ AL Q + + ++ + + T
Sbjct 472 RDKEMEKSFEVVRHKNRDREEVSKNQALKLQLDHQYAMLENQKYSHT 518
> hsa:84694 GJA10, CX62, RP11-63K6.6; gap junction protein, alpha
10, 62kDa; K07631 connexin 62
Length=543
Score = 28.5 bits (62), Expect = 8.4, Method: Composition-based stats.
Identities = 26/99 (26%), Positives = 42/99 (42%), Gaps = 11/99 (11%)
Query 53 KIVKPRNPRKLLCKHFLVGKTKGEKGEQQKATDALAQQSNPDLGLDDDNRATATLEVDAG 112
+I+ +P + H L EK Q+K + AQ NPDL L++ R L
Sbjct 81 QIIFVSSPSLVYMGHALYRLRAFEKDRQRKKSHLRAQMENPDLDLEEQQRIDRELR---- 136
Query 113 RGEDKRALWE-------RRLAISETLKKGLLEPGVYRGQ 144
R E+++ + + R + L + +LE G GQ
Sbjct 137 RLEEQKRIHKVPLKGCLLRTYVLHILTRSVLEVGFMIGQ 175
Lambda K H
0.312 0.134 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3256415000
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40