bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7714_orf1
Length=116
Score E
Sequences producing significant alignments: (Bits) Value
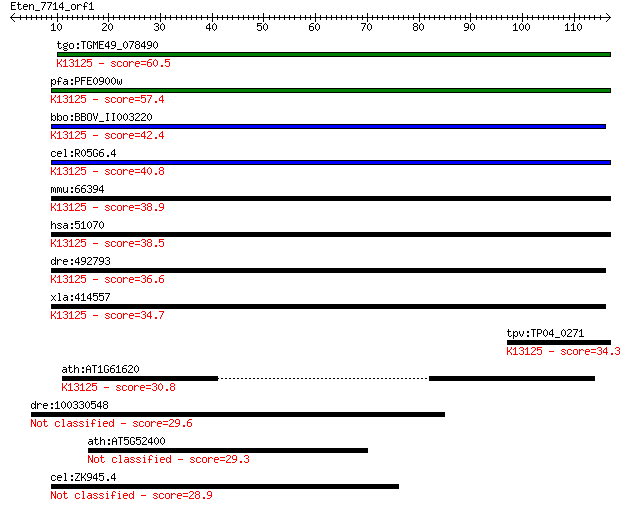
tgo:TGME49_078490 hypothetical protein ; K13125 nitric oxide s... 60.5 1e-09
pfa:PFE0900w conserved protein, unknown function; K13125 nitri... 57.4 1e-08
bbo:BBOV_II003220 18.m06270; hypothetical protein; K13125 nitr... 42.4 4e-04
cel:R05G6.4 hypothetical protein; K13125 nitric oxide synthase... 40.8 0.001
mmu:66394 Nosip, 2310061K06Rik, CGI-25, MGC115777; nitric oxid... 38.9 0.004
hsa:51070 NOSIP; nitric oxide synthase interacting protein; K1... 38.5 0.005
dre:492793 nosip, wu:fb80a11, zgc:101669; nitric oxide synthas... 36.6 0.020
xla:414557 nosip, MGC78783; nitric oxide synthase interacting ... 34.7 0.073
tpv:TP04_0271 hypothetical protein; K13125 nitric oxide syntha... 34.3 0.097
ath:AT1G61620 phosphoinositide binding; K13125 nitric oxide sy... 30.8 1.1
dre:100330548 novel protein with a Immunoglobulin V-set domain 29.6 2.7
ath:AT5G52400 CYP715A1; electron carrier/ heme binding / iron ... 29.3 3.2
cel:ZK945.4 hypothetical protein 28.9 4.1
> tgo:TGME49_078490 hypothetical protein ; K13125 nitric oxide
synthase-interacting protein
Length=322
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 39/107 (36%), Positives = 44/107 (41%), Gaps = 51/107 (47%)
Query 10 ARHSKNATSAPFYSYHERKKIRGQTRVENFGSSCSSYMEHRVAQLRLPQFRRFQMSRNEV 69
ARHSKNATSA FYSYHERKK++
Sbjct 3 ARHSKNATSAAFYSYHERKKLK-------------------------------------- 24
Query 70 MSLSLFLFAFAALGVGTLKGRLDTRACRRFESCWLCLSPARNPLATP 116
VGT + RLDT A RRFE+CWLC A P+ TP
Sbjct 25 -------------DVGTQRERLDTDALRRFEACWLCNRTALAPVCTP 58
> pfa:PFE0900w conserved protein, unknown function; K13125 nitric
oxide synthase-interacting protein
Length=270
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 32/108 (29%), Positives = 44/108 (40%), Gaps = 51/108 (47%)
Query 9 MARHSKNATSAPFYSYHERKKIRGQTRVENFGSSCSSYMEHRVAQLRLPQFRRFQMSRNE 68
MARHSKN T+ P ++YHERKK++
Sbjct 1 MARHSKNNTANPIFTYHERKKVKD------------------------------------ 24
Query 69 VMSLSLFLFAFAALGVGTLKGRLDTRACRRFESCWLCLSPARNPLATP 116
VGTL+ RL + R+FE CW+CL A NP+++P
Sbjct 25 ---------------VGTLRERLGKDSMRKFEQCWICLRTAENPVSSP 57
> bbo:BBOV_II003220 18.m06270; hypothetical protein; K13125 nitric
oxide synthase-interacting protein
Length=262
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 28/107 (26%), Positives = 37/107 (34%), Gaps = 51/107 (47%)
Query 9 MARHSKNATSAPFYSYHERKKIRGQTRVENFGSSCSSYMEHRVAQLRLPQFRRFQMSRNE 68
M RHSKN T+ P ++YHERK ++
Sbjct 1 MTRHSKNNTANPIFTYHERKNVK------------------------------------- 23
Query 69 VMSLSLFLFAFAALGVGTLKGRLDTRACRRFESCWLCLSPARNPLAT 115
T + RL + RR E CWLCLS A P++T
Sbjct 24 --------------DFNTQRQRLGADSMRRCEQCWLCLSTAEKPVST 56
> cel:R05G6.4 hypothetical protein; K13125 nitric oxide synthase-interacting
protein
Length=310
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 30/108 (27%), Positives = 39/108 (36%), Gaps = 48/108 (44%)
Query 9 MARHSKNATSAPFYSYHERKKIRGQTRVENFGSSCSSYMEHRVAQLRLPQFRRFQMSRNE 68
M RH KN+T+A Y+YHER+ R A+
Sbjct 1 MTRHGKNSTAASVYTYHERR---------------------RDAK--------------- 24
Query 69 VMSLSLFLFAFAALGVGTLKGRLDTRACRRFESCWLCLSPARNPLATP 116
A G GTL RL + + F C L L P RNP+ +P
Sbjct 25 ------------ASGYGTLHARLGADSIKEFHCCSLTLQPCRNPVISP 60
> mmu:66394 Nosip, 2310061K06Rik, CGI-25, MGC115777; nitric oxide
synthase interacting protein; K13125 nitric oxide synthase-interacting
protein
Length=276
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 28/108 (25%), Positives = 35/108 (32%), Gaps = 48/108 (44%)
Query 9 MARHSKNATSAPFYSYHERKKIRGQTRVENFGSSCSSYMEHRVAQLRLPQFRRFQMSRNE 68
M RH KN T+ Y+YHE+KK
Sbjct 1 MTRHGKNCTAGAVYTYHEKKK--------------------------------------- 21
Query 69 VMSLSLFLFAFAALGVGTLKGRLDTRACRRFESCWLCLSPARNPLATP 116
AA G GT RL A + F+ C L L P +P+ TP
Sbjct 22 ---------DTAASGYGTQNIRLSRDAVKDFDCCCLSLQPCHDPVVTP 60
> hsa:51070 NOSIP; nitric oxide synthase interacting protein;
K13125 nitric oxide synthase-interacting protein
Length=301
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 28/108 (25%), Positives = 35/108 (32%), Gaps = 48/108 (44%)
Query 9 MARHSKNATSAPFYSYHERKKIRGQTRVENFGSSCSSYMEHRVAQLRLPQFRRFQMSRNE 68
M RH KN T+ Y+YHE+KK
Sbjct 1 MTRHGKNCTAGAVYTYHEKKK--------------------------------------- 21
Query 69 VMSLSLFLFAFAALGVGTLKGRLDTRACRRFESCWLCLSPARNPLATP 116
AA G GT RL A + F+ C L L P +P+ TP
Sbjct 22 ---------DTAASGYGTQNIRLSRDAVKDFDCCCLSLQPCHDPVVTP 60
> dre:492793 nosip, wu:fb80a11, zgc:101669; nitric oxide synthase
interacting protein; K13125 nitric oxide synthase-interacting
protein
Length=304
Score = 36.6 bits (83), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 27/107 (25%), Positives = 35/107 (32%), Gaps = 48/107 (44%)
Query 9 MARHSKNATSAPFYSYHERKKIRGQTRVENFGSSCSSYMEHRVAQLRLPQFRRFQMSRNE 68
M RH KN T+ Y+YHE++K
Sbjct 1 MTRHGKNCTAGAVYTYHEKRK--------------------------------------- 21
Query 69 VMSLSLFLFAFAALGVGTLKGRLDTRACRRFESCWLCLSPARNPLAT 115
AA G GT RL A + F+ C L L P R+P+ T
Sbjct 22 ---------DTAASGYGTQSVRLGKDAIKDFDCCSLSLQPCRDPVLT 59
> xla:414557 nosip, MGC78783; nitric oxide synthase interacting
protein; K13125 nitric oxide synthase-interacting protein
Length=298
Score = 34.7 bits (78), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 25/107 (23%), Positives = 34/107 (31%), Gaps = 48/107 (44%)
Query 9 MARHSKNATSAPFYSYHERKKIRGQTRVENFGSSCSSYMEHRVAQLRLPQFRRFQMSRNE 68
M RH KN T+ Y+Y+E+KK
Sbjct 1 MTRHGKNCTAGAVYTYYEKKK--------------------------------------- 21
Query 69 VMSLSLFLFAFAALGVGTLKGRLDTRACRRFESCWLCLSPARNPLAT 115
A G GT RL A + F+ C L L P ++P+ T
Sbjct 22 ---------DTVASGYGTQTVRLSKDAVKDFDCCCLSLQPCKDPVVT 59
> tpv:TP04_0271 hypothetical protein; K13125 nitric oxide synthase-interacting
protein
Length=241
Score = 34.3 bits (77), Expect = 0.097, Method: Compositional matrix adjust.
Identities = 12/20 (60%), Positives = 15/20 (75%), Gaps = 0/20 (0%)
Query 97 RRFESCWLCLSPARNPLATP 116
R+FE CWLCL+ A P+ TP
Sbjct 2 RKFEQCWLCLATAVKPVTTP 21
> ath:AT1G61620 phosphoinositide binding; K13125 nitric oxide
synthase-interacting protein
Length=310
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 82 LGVGTLKGRLDTRACRRFESCWLCLSPARNPL 113
LG GT + RL + + F++C LCL P +P+
Sbjct 23 LGYGTQRERLGRDSIKPFDACSLCLKPFIDPM 54
Score = 28.5 bits (62), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 12/30 (40%), Positives = 18/30 (60%), Gaps = 0/30 (0%)
Query 11 RHSKNATSAPFYSYHERKKIRGQTRVENFG 40
RHSKN +++Y E+KK+ T+ E G
Sbjct 4 RHSKNNNDLAYFTYDEKKKLGYGTQRERLG 33
> dre:100330548 novel protein with a Immunoglobulin V-set domain
Length=523
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 21/96 (21%), Positives = 42/96 (43%), Gaps = 16/96 (16%)
Query 5 ENRAMARHSKNATSAPFYSYHERKKIR-------GQTRVENFGSSCSSYMEHRVAQLRLP 57
E + +A+ K S+P+Y+ ER K R G ++ + + Y R++ +
Sbjct 239 EGKLLAKEDKETKSSPYYNADERYKGRLQLNDQTGFLTLDKMKDTDAGYYTVRISSNKQT 298
Query 58 QFRRFQM---------SRNEVMSLSLFLFAFAALGV 84
+++F + S +++L L + A LGV
Sbjct 299 TYKKFAVNFTGSVVSPSAGAIIALLLVIAVVAGLGV 334
> ath:AT5G52400 CYP715A1; electron carrier/ heme binding / iron
ion binding / monooxygenase/ oxygen binding
Length=519
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 16/54 (29%), Positives = 29/54 (53%), Gaps = 1/54 (1%)
Query 16 ATSAPFYSYHERKKIRGQTRVENFGSSCSSYMEHRVAQLRLPQFRRFQMSRNEV 69
A S PF + ++ KK++ + V + S S+ + H + + LP F R+Q +V
Sbjct 52 APSFPFGNLNDMKKLKMASVVVD-NSKSSTIINHDIHSIALPHFARWQQEYGKV 104
> cel:ZK945.4 hypothetical protein
Length=499
Score = 28.9 bits (63), Expect = 4.1, Method: Composition-based stats.
Identities = 25/82 (30%), Positives = 34/82 (41%), Gaps = 20/82 (24%)
Query 9 MARHSKNATSAPFYSYHERKKI----------RGQTRV---ENFGSSCSSYMEHRVA--Q 53
+A H K S P +H K I + Q R+ E F CS + EH +
Sbjct 139 LAHHEKKMISTPDCQFHPGKSISLVCMRDRCKKRQNRLMCSECFLDKCSDHFEHEHSSLH 198
Query 54 LRLPQFRRFQMSRNEVMSLSLF 75
L LP+ R RN SL+L+
Sbjct 199 LELPELR-----RNICSSLALY 215
Lambda K H
0.325 0.132 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2037741960
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40