bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7759_orf1
Length=110
Score E
Sequences producing significant alignments: (Bits) Value
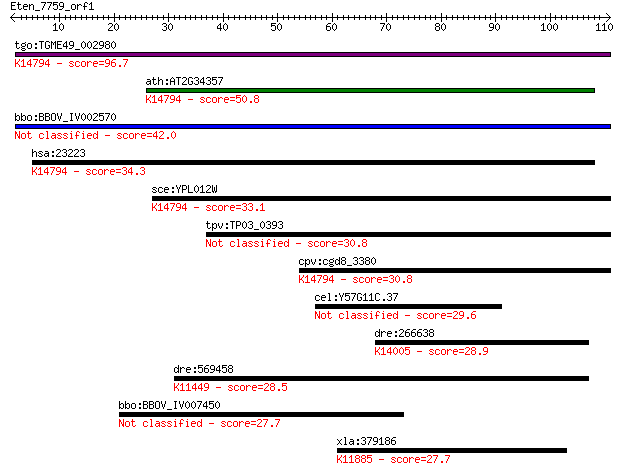
tgo:TGME49_002980 hypothetical protein ; K14794 ribosomal RNA-... 96.7 2e-20
ath:AT2G34357 binding; K14794 ribosomal RNA-processing protein 12 50.8
bbo:BBOV_IV002570 21.m02817; hypothetical protein 42.0 5e-04
hsa:23223 RRP12, DKFZp762P1116, FLJ20231, KIAA0690; ribosomal ... 34.3 0.098
sce:YPL012W RRP12; Protein required for export of the ribosoma... 33.1 0.22
tpv:TP03_0393 hypothetical protein 30.8 1.0
cpv:cgd8_3380 Rrp12p like nucleolar protein ; K14794 ribosomal... 30.8 1.2
cel:Y57G11C.37 hypothetical protein 29.6 2.7
dre:266638 sec31a, cb354, sec31p, zgc:63547; sec31 homolog A (... 28.9 4.1
dre:569458 im:7163548; K11449 jumonji domain-containing protei... 28.5 6.2
bbo:BBOV_IV007450 23.m05961; ribosomal protein L2 27.7 8.4
xla:379186 ddi2, MGC53726; protein DDI1 homolog 2; K11885 DNA ... 27.7 9.3
> tgo:TGME49_002980 hypothetical protein ; K14794 ribosomal RNA-processing
protein 12
Length=1915
Score = 96.7 bits (239), Expect = 2e-20, Method: Composition-based stats.
Identities = 50/109 (45%), Positives = 72/109 (66%), Gaps = 0/109 (0%)
Query 2 LGRLFDLYCGELPHSLVQQWAAVGLLLLKDHNKIAFVAGIQFAKACIRAGDMDDLAAWLP 61
L R+ Y ++ +L++Q A V LLLL+D +K F+A ++FA+ D L +LP
Sbjct 1436 LSRVIFSYADQMDGALIKQIAEVVLLLLQDRDKQVFLAALRFARVIAHVFDGQALERFLP 1495
Query 62 SMLGVLKNPNAPKARMKIRHLIEKLCKVMGEEAVEKAMPEEHMPLLRHL 110
+ML L N +A +A+MKIR L+EKL K +GE AV++A P EH+PLLRHL
Sbjct 1496 AMLCALNNKHAIRAKMKIRRLVEKLLKKLGEAAVKEAFPSEHLPLLRHL 1544
> ath:AT2G34357 binding; K14794 ribosomal RNA-processing protein
12
Length=1280
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 35/85 (41%), Positives = 46/85 (54%), Gaps = 3/85 (3%)
Query 26 LLLLKDHNKIAFVAGIQFAKACIRAGDMDDLAAWLPSML-GVLKNPNAPKA--RMKIRHL 82
LLL+ NK A + K + ++ L A L SM+ G+LK P K + K+R L
Sbjct 923 FLLLQRKNKEITKANLGLLKVLVAKSPVEGLHANLKSMVEGLLKWPEGTKNLFKAKVRLL 982
Query 83 IEKLCKVMGEEAVEKAMPEEHMPLL 107
+E L K G EAV+ MPEEHM LL
Sbjct 983 LEMLIKKCGTEAVKSVMPEEHMKLL 1007
> bbo:BBOV_IV002570 21.m02817; hypothetical protein
Length=1281
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 26/111 (23%), Positives = 58/111 (52%), Gaps = 3/111 (2%)
Query 2 LGRLFDLYCGE-LPHSLVQQWAAVGLLLLKDHNKIAFVAGIQFAKACIRAGDMDDLAAWL 60
L RL + E + ++L Q ++ V LL K+ ++ ++FA+ CI + + +
Sbjct 1056 LTRLMARHSSEAVKYNLTQVFSYVFRLLSMADQKL-YIQLLKFARVCIVKLNQEQVLWLA 1114
Query 61 PSMLGVLKNPNA-PKARMKIRHLIEKLCKVMGEEAVEKAMPEEHMPLLRHL 110
P ++ + N ++++ +R +++KL + + + P+EH+PL+ HL
Sbjct 1115 PMLMKMFDNATCCQRSKVYVRRVVQKLMVKLSRDQIIHIFPQEHLPLMHHL 1165
> hsa:23223 RRP12, DKFZp762P1116, FLJ20231, KIAA0690; ribosomal
RNA processing 12 homolog (S. cerevisiae); K14794 ribosomal
RNA-processing protein 12
Length=1236
Score = 34.3 bits (77), Expect = 0.098, Method: Compositional matrix adjust.
Identities = 29/106 (27%), Positives = 49/106 (46%), Gaps = 4/106 (3%)
Query 5 LFDLYCGELPHSLVQQWAAVGLLLLKDHNKIAFVAGIQFAKACIRAGDMDDLAAWLP--- 61
LF+ + G + S V+Q LLL + + + F K + D+ LA +
Sbjct 867 LFE-FKGLMGTSTVEQLLENVCLLLASRTRDVVKSALGFIKVAVTVMDVAHLAKHVQLVM 925
Query 62 SMLGVLKNPNAPKARMKIRHLIEKLCKVMGEEAVEKAMPEEHMPLL 107
+G L + RMK+R+L K + G E V++ +PEE+ +L
Sbjct 926 EAIGKLSDDMRRHFRMKLRNLFTKFIRKFGFELVKRLLPEEYHRVL 971
> sce:YPL012W RRP12; Protein required for export of the ribosomal
subunits; associates with the RNA components of the pre-ribosomes;
contains HEAT-repeats; K14794 ribosomal RNA-processing
protein 12
Length=1228
Score = 33.1 bits (74), Expect = 0.22, Method: Composition-based stats.
Identities = 21/87 (24%), Positives = 42/87 (48%), Gaps = 3/87 (3%)
Query 27 LLLKDHNKIAFVAGIQFAKACIRAGDMDDLAAWLPSMLGVL---KNPNAPKARMKIRHLI 83
L L +++ + I F K C+ + + +P +L L + + + K++H+I
Sbjct 933 LYLTSNSREIVKSAIGFTKVCVLGLPEELMRPKVPELLLKLLRWSHEHTGHFKAKVKHII 992
Query 84 EKLCKVMGEEAVEKAMPEEHMPLLRHL 110
E+L + G + +E PEE LL ++
Sbjct 993 ERLIRRFGYDYIEANFPEEDRRLLTNI 1019
> tpv:TP03_0393 hypothetical protein
Length=1424
Score = 30.8 bits (68), Expect = 1.0, Method: Composition-based stats.
Identities = 15/75 (20%), Positives = 39/75 (52%), Gaps = 1/75 (1%)
Query 37 FVAGIQFAKACIRAGDMDDLAAWLPSMLGVLKNPNA-PKARMKIRHLIEKLCKVMGEEAV 95
++ ++F + + + D+L P+++ + N KA++ +R L+EK+ + + +
Sbjct 1247 YIQILKFFRIVMVNMNHDNLVKVTPNIMKLFNNERCCSKAKIYVRRLVEKMLTRLKRDYL 1306
Query 96 EKAMPEEHMPLLRHL 110
P+EH L+ ++
Sbjct 1307 FSIFPKEHFSLVTNI 1321
> cpv:cgd8_3380 Rrp12p like nucleolar protein ; K14794 ribosomal
RNA-processing protein 12
Length=1339
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 17/58 (29%), Positives = 30/58 (51%), Gaps = 1/58 (1%)
Query 54 DDLAAWLPSMLG-VLKNPNAPKARMKIRHLIEKLCKVMGEEAVEKAMPEEHMPLLRHL 110
D + + P+++ +L N + K R+ +R ++ L K G E + P EH L R+L
Sbjct 1029 DLMWNYTPNIISNILNNQCSLKFRIYVRKILISLIKRFGAEKTSQVFPLEHNQLYRYL 1086
> cel:Y57G11C.37 hypothetical protein
Length=837
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 14/41 (34%), Positives = 21/41 (51%), Gaps = 7/41 (17%)
Query 57 AAWLPSMLGVL-------KNPNAPKARMKIRHLIEKLCKVM 90
A W PS+LG+L K N+P + + LI +C V+
Sbjct 239 ALWFPSLLGILMWFLGGFKYKNSPGDKQDLYQLISDICFVL 279
> dre:266638 sec31a, cb354, sec31p, zgc:63547; sec31 homolog A
(S. cerevisiae); K14005 protein transport protein SEC31
Length=1254
Score = 28.9 bits (63), Expect = 4.1, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 25/45 (55%), Gaps = 6/45 (13%)
Query 68 KNPNAPKARMK------IRHLIEKLCKVMGEEAVEKAMPEEHMPL 106
+NP+ P+ M+ I +I+ L + E+ +K +PEEHM L
Sbjct 1109 RNPSMPQGNMEGAPGAPIGDVIKPLQAIPTEKITKKPIPEEHMVL 1153
> dre:569458 im:7163548; K11449 jumonji domain-containing protein
1 [EC:1.14.11.-]
Length=1581
Score = 28.5 bits (62), Expect = 6.2, Method: Composition-based stats.
Identities = 23/80 (28%), Positives = 35/80 (43%), Gaps = 9/80 (11%)
Query 31 DHNKIAFVAGIQFAKACIRAGDMDDLAAW----LPSMLGVLKNPNAPKARMKIRHLIEKL 86
D A +AG + I GD+DD+ + G L + A K KIR L+ K+
Sbjct 1393 DQESEADLAGFKEVMQTIEEGDVDDMTKRRVYEIKEKPGALWHIYAAKDAEKIRELLRKV 1452
Query 87 CKVMGEEAVEKAMPEEHMPL 106
+ G+E P +H P+
Sbjct 1453 GEEQGQEN-----PPDHDPI 1467
> bbo:BBOV_IV007450 23.m05961; ribosomal protein L2
Length=332
Score = 27.7 bits (60), Expect = 8.4, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 25/52 (48%), Gaps = 11/52 (21%)
Query 21 WAAVGLLLLKDHNKIAFVAGIQFAKACIRAGDMDDLAAWLPSMLGVLKNPNA 72
WA +G + +HNK + +AG +L W PS+ G+ NPNA
Sbjct 234 WATLGQVSNIEHNK----------RILKKAGTRRNLG-WRPSVRGIAMNPNA 274
> xla:379186 ddi2, MGC53726; protein DDI1 homolog 2; K11885 DNA
damage-inducible protein 1
Length=393
Score = 27.7 bits (60), Expect = 9.3, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 22/42 (52%), Gaps = 0/42 (0%)
Query 61 PSMLGVLKNPNAPKARMKIRHLIEKLCKVMGEEAVEKAMPEE 102
P L +LK N P A + +EK KV+ E+ E+A E+
Sbjct 140 PHELSLLKERNPPLAEALLSGDLEKFTKVLQEQQQERARREQ 181
Lambda K H
0.325 0.140 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2067351240
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40