bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7869_orf1
Length=177
Score E
Sequences producing significant alignments: (Bits) Value
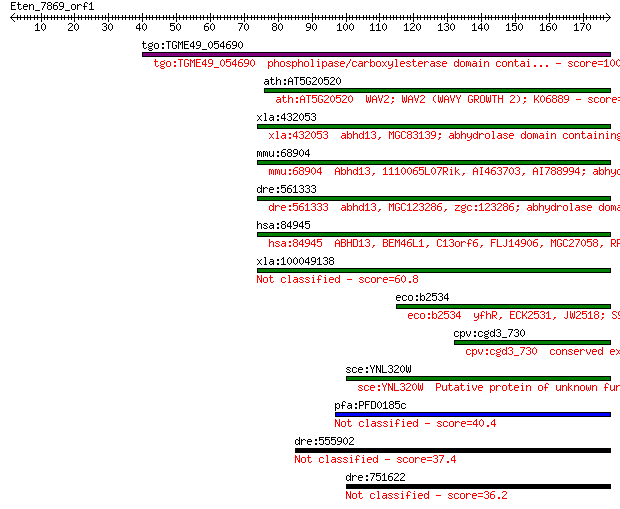
tgo:TGME49_054690 phospholipase/carboxylesterase domain contai... 100 2e-21
ath:AT5G20520 WAV2; WAV2 (WAVY GROWTH 2); K06889 73.6 3e-13
xla:432053 abhd13, MGC83139; abhydrolase domain containing 13;... 72.8 6e-13
mmu:68904 Abhd13, 1110065L07Rik, AI463703, AI788994; abhydrola... 70.9 2e-12
dre:561333 abhd13, MGC123286, zgc:123286; abhydrolase domain c... 69.7 5e-12
hsa:84945 ABHD13, BEM46L1, C13orf6, FLJ14906, MGC27058, RP11-1... 61.2 2e-09
xla:100049138 hypothetical protein LOC100049138 60.8 2e-09
eco:b2534 yfhR, ECK2531, JW2518; S9 peptidase family protein, ... 53.9 3e-07
cpv:cgd3_730 conserved expressed protein ; K06889 52.4 8e-07
sce:YNL320W Putative protein of unknown function; the authenti... 51.2 2e-06
pfa:PFD0185c conserved Plasmodium protein, unknown function 40.4 0.003
dre:555902 Bem46-like 37.4 0.030
dre:751622 MGC153037, zgc:153037; si:ch211-117n7.7 36.2 0.064
> tgo:TGME49_054690 phospholipase/carboxylesterase domain containing
protein (EC:3.1.-.-); K06889
Length=497
Score = 100 bits (250), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 60/142 (42%), Positives = 76/142 (53%), Gaps = 4/142 (2%)
Query 40 AAEPAAQGAAAAAAAAAAAAAAAAAAATKRHDSEFSFQEKLVFDPSYPIEPAEIKKFPHI 99
AA PAA A A AA A + FQEKL+F P P K P
Sbjct 11 AANPAASIFLAIAGFAARATWVGGLILLCMVVLLWYFQEKLLFYPGVPQGFETPDKNPKG 70
Query 100 IGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTENE---KAPTFILFHGNYGHVGLTLPRA 156
+ SPA G+P+++L+L T DGV++H W +KQ + APT I FHGN G+VG LP
Sbjct 71 LRSPAERGLPFEELWLRTVDGVKLHCWLIKQKLPQVAAHAPTLIFFHGNAGNVGFRLPNV 130
Query 157 RWLYDQ-GMNVLVIDYRGYGRS 177
LY G+NVL++ YRGYG S
Sbjct 131 ELLYKHVGVNVLIVSYRGYGFS 152
> ath:AT5G20520 WAV2; WAV2 (WAVY GROWTH 2); K06889
Length=308
Score = 73.6 bits (179), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 38/103 (36%), Positives = 57/103 (55%), Gaps = 7/103 (6%)
Query 76 FQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTENEK 135
FQEKLV+ P+ P K +P +PA + Y+D++L + DGVR+H WF+K +
Sbjct 26 FQEKLVY---VPVLPGLSKSYPI---TPARLNLIYEDIWLQSSDGVRLHAWFIKMFPECR 79
Query 136 APTFILFHGNYGHVGLTLPRARWLYDQ-GMNVLVIDYRGYGRS 177
PT + F N G++ L R + + NV ++ YRGYG S
Sbjct 80 GPTILFFQENAGNIAHRLEMVRIMIQKLKCNVFMLSYRGYGAS 122
> xla:432053 abhd13, MGC83139; abhydrolase domain containing 13;
K06889
Length=336
Score = 72.8 bits (177), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 38/107 (35%), Positives = 66/107 (61%), Gaps = 11/107 (10%)
Query 74 FSFQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTEN 133
F FQ+ L++ +P +P+ + + P GIP++++F+ TKD +R++ L+ T +
Sbjct 58 FKFQDVLLY---FPDQPSSSRLY-----IPMPTGIPHENIFIKTKDNIRLNLILLRYTGD 109
Query 134 EKA--PTFILFHGNYGHVGLTLPRA-RWLYDQGMNVLVIDYRGYGRS 177
+ PT I FHGN G++G LP A L + +N++++DYRGYG+S
Sbjct 110 NSSFSPTIIYFHGNAGNIGHRLPNALLMLVNLKVNLILVDYRGYGKS 156
> mmu:68904 Abhd13, 1110065L07Rik, AI463703, AI788994; abhydrolase
domain containing 13; K06889
Length=337
Score = 70.9 bits (172), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 38/107 (35%), Positives = 66/107 (61%), Gaps = 11/107 (10%)
Query 74 FSFQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTEN 133
+ FQ+ L++ +P +P+ + + P GIP++++F+ TKDGVR++ ++ T +
Sbjct 58 YKFQDVLLY---FPEQPSSSRLY-----VPMPTGIPHENIFIRTKDGVRLNLILVRYTGD 109
Query 134 EK--APTFILFHGNYGHVGLTLPRA-RWLYDQGMNVLVIDYRGYGRS 177
PT I FHGN G++G LP A L + +N++++DYRGYG+S
Sbjct 110 NSPYCPTIIYFHGNAGNIGHRLPNALLMLVNLRVNLVLVDYRGYGKS 156
> dre:561333 abhd13, MGC123286, zgc:123286; abhydrolase domain
containing 13; K06889
Length=337
Score = 69.7 bits (169), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 39/107 (36%), Positives = 67/107 (62%), Gaps = 11/107 (10%)
Query 74 FSFQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQT-E 132
+ FQ+ L++ +P +P+ + + P GIP++++++ TKDG+R++ L+ T E
Sbjct 58 YKFQDVLLY---FPDQPSSSRLY-----VPMPTGIPHENVYIRTKDGIRLNLILLRYTGE 109
Query 133 NE-KAPTFILFHGNYGHVGLTLPRA-RWLYDQGMNVLVIDYRGYGRS 177
N APT + FHGN G++G +P A L + NV+++DYRGYG+S
Sbjct 110 NPAGAPTILYFHGNAGNIGHRVPNALLMLVNLKANVVLVDYRGYGKS 156
> hsa:84945 ABHD13, BEM46L1, C13orf6, FLJ14906, MGC27058, RP11-153I24.2,
bA153I24.2; abhydrolase domain containing 13; K06889
Length=337
Score = 61.2 bits (147), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 37/107 (34%), Positives = 67/107 (62%), Gaps = 11/107 (10%)
Query 74 FSFQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTEN 133
+ FQ+ L++ +P +P+ + + P GIP++++F+ TKDG+R++ ++ T +
Sbjct 58 YKFQDVLLY---FPEQPSSSRLY-----VPMPTGIPHENIFIRTKDGIRLNLILIRYTGD 109
Query 134 EK--APTFILFHGNYGHVGLTLPRARWL-YDQGMNVLVIDYRGYGRS 177
+PT I FHGN G++G LP A + + +N+L++DYRGYG+S
Sbjct 110 NSPYSPTIIYFHGNAGNIGHRLPNALLMLVNLKVNLLLVDYRGYGKS 156
> xla:100049138 hypothetical protein LOC100049138
Length=336
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 37/107 (34%), Positives = 66/107 (61%), Gaps = 11/107 (10%)
Query 74 FSFQEKLVFDPSYPIEPAEIKKFPHIIGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTEN 133
F FQ+ L++ +P +P+ + + P GIP++++F+ TKD +R++ L+ T +
Sbjct 58 FKFQDVLLY---FPDQPSSSRLY-----IPMPTGIPHENIFIKTKDNIRLNLILLRYTGD 109
Query 134 EK--APTFILFHGNYGHVGLTLPRARWL-YDQGMNVLVIDYRGYGRS 177
+PT + FHGN G++G LP A + + +N+L++DYRGYG+S
Sbjct 110 NSNFSPTIVYFHGNAGNIGHRLPNALLMLVNLKVNLLLVDYRGYGKS 156
> eco:b2534 yfhR, ECK2531, JW2518; S9 peptidase family protein,
function unknown; K06889
Length=284
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 25/66 (37%), Positives = 36/66 (54%), Gaps = 3/66 (4%)
Query 115 LTTKDGVRIHGWFLKQTE---NEKAPTFILFHGNYGHVGLTLPRARWLYDQGMNVLVIDY 171
T KDG R+ GWF+ + + T I HGN G++ P WL ++ NV + DY
Sbjct 54 FTAKDGTRLQGWFIPSSTGPADNAIATIIHAHGNAGNMSAHWPLVSWLPERNFNVFMFDY 113
Query 172 RGYGRS 177
RG+G+S
Sbjct 114 RGFGKS 119
> cpv:cgd3_730 conserved expressed protein ; K06889
Length=419
Score = 52.4 bits (124), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 23/47 (48%), Positives = 31/47 (65%), Gaps = 1/47 (2%)
Query 132 ENEKAPTFILFHGNYGHVGLTLPRARWLYDQ-GMNVLVIDYRGYGRS 177
+ EKAPT + FHGN G++G LPR Y+ G+N+ + YRGYG S
Sbjct 160 QQEKAPTIVFFHGNAGNIGHRLPRFLEFYNLIGVNIFAVSYRGYGDS 206
> sce:YNL320W Putative protein of unknown function; the authentic,
non-tagged protein is detected in highly purified mitochondria
in high-throughput studies; K06889
Length=284
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 30/79 (37%), Positives = 44/79 (55%), Gaps = 3/79 (3%)
Query 100 IGSPASYGIPYDDLFLTTKDGVRIHGWFLKQTENEKAPTFILFHGNYGHVGLTLPRARWL 159
+ +P S GIPY+ L L T+D +++ W +K EN + IL N G++G +
Sbjct 44 VDTPDSRGIPYEKLTLITQDHIKLEAWDIKN-ENSTSTVLIL-CPNAGNIGYFILIIDIF 101
Query 160 YDQ-GMNVLVIDYRGYGRS 177
Y Q GM+V + YRGYG S
Sbjct 102 YRQFGMSVFIYSYRGYGNS 120
> pfa:PFD0185c conserved Plasmodium protein, unknown function
Length=734
Score = 40.4 bits (93), Expect = 0.003, Method: Composition-based stats.
Identities = 33/84 (39%), Positives = 41/84 (48%), Gaps = 11/84 (13%)
Query 97 PHIIGSPASYGIPYDDL-FLTTKDGVRIHGWFLKQTENEKAPTFILF-HGNYGHVGLTLP 154
PH P SY +L F+ TK G I G FL N A ILF HGN +G +P
Sbjct 12 PH----PPSYSKNRKNLHFIKTKHGSTICGIFL----NNNAHLTILFSHGNAEDIGDIVP 63
Query 155 R-ARWLYDQGMNVLVIDYRGYGRS 177
+ L G+N+ DY GYG+S
Sbjct 64 QFESKLKRLGLNMFAYDYSGYGQS 87
> dre:555902 Bem46-like
Length=344
Score = 37.4 bits (85), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 38/120 (31%), Positives = 53/120 (44%), Gaps = 28/120 (23%)
Query 85 SYPIEPAEIKKFPHIIGSPASYGIPYD-------DLFLTTKDGVRI--------HGWFLK 129
S+ + A I H++ P D +++L ++GVR+ H W
Sbjct 42 SFLVLCATIPTLQHVLSDFVDLSHPLDVGLNHTINVYLKPEEGVRVGVWHTVPEHRWKEA 101
Query 130 QTEN----EKA-----PTFILFHGNYGHVGLTLPR---ARWLYDQGMNVLVIDYRGYGRS 177
Q +N EKA P FI HGN G+ L R A L G +VLV+DYRG+G S
Sbjct 102 QGKNAEWYEKALGDGSPIFIYLHGNGGNRS-ALHRIGVANVLSALGYHVLVMDYRGFGDS 160
> dre:751622 MGC153037, zgc:153037; si:ch211-117n7.7
Length=347
Score = 36.2 bits (82), Expect = 0.064, Method: Compositional matrix adjust.
Identities = 32/102 (31%), Positives = 49/102 (48%), Gaps = 28/102 (27%)
Query 100 IGSPASYGIPYD-DLFLTTKDGVRI--------HGWFLKQTEN----EKA-----PTFIL 141
+ P+ G+ + + +L T++GVR+ H W Q +N EKA P F+
Sbjct 65 LSRPSDLGLNHTINFYLKTEEGVRVGVWHTVPEHRWKEAQGKNVEWYEKALGDGSPIFMY 124
Query 142 FHGNYGH------VGLTLPRARWLYDQGMNVLVIDYRGYGRS 177
HGN G+ +G+ A L G + LV+DYRG+G S
Sbjct 125 LHGNTGNRSAPHRIGV----ANILSALGYHALVMDYRGFGDS 162
Lambda K H
0.317 0.128 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4665550176
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40