bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7908_orf1
Length=178
Score E
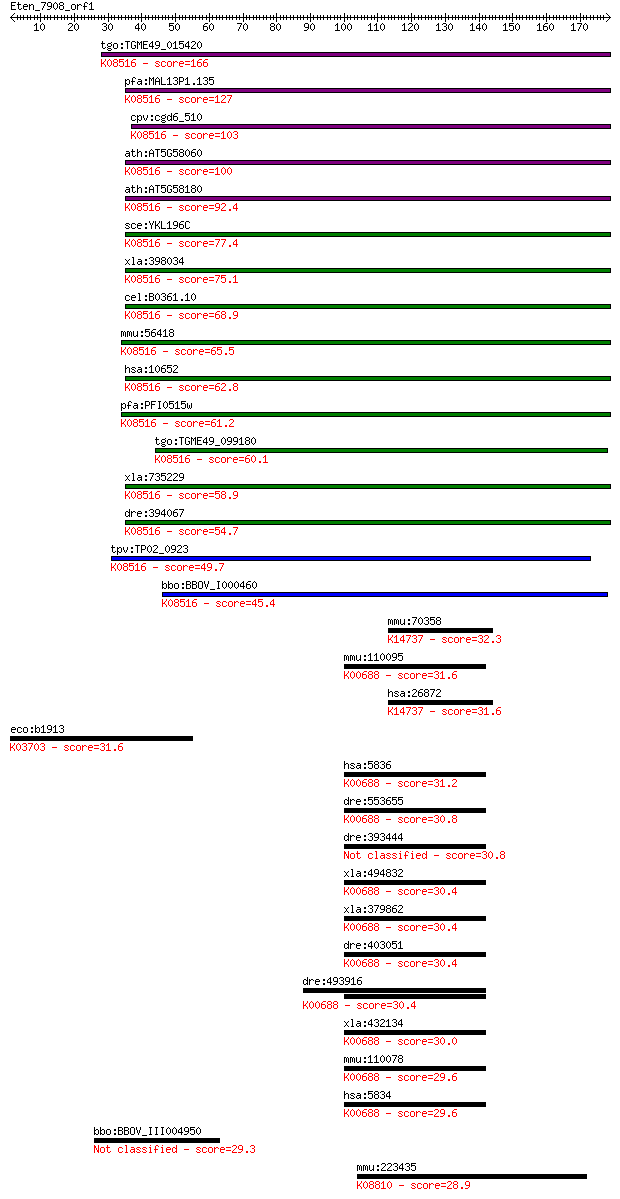
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_015420 SNARE protein, putative ; K08516 synaptobrev... 166 4e-41
pfa:MAL13P1.135 PfYkt6.2; SNARE protein, putative; K08516 syna... 127 1e-29
cpv:cgd6_510 synaptobrevin/VAMP-like protein ; K08516 synaptob... 103 3e-22
ath:AT5G58060 YKT61; YKT61; K08516 synaptobrevin homolog YKT6 100 3e-21
ath:AT5G58180 ATYKT62; K08516 synaptobrevin homolog YKT6 92.4 7e-19
sce:YKL196C YKT6; Vesicle membrane protein (v-SNARE) with acyl... 77.4 2e-14
xla:398034 ykt6-a, ykt6, ykt6a; YKT6 v-SNARE homolog (EC:2.3.1... 75.1 1e-13
cel:B0361.10 hypothetical protein; K08516 synaptobrevin homolo... 68.9 8e-12
mmu:56418 Ykt6, 0610042I15Rik, 1810013M05Rik, AW105923; YKT6 h... 65.5 9e-11
hsa:10652 YKT6; YKT6 v-SNARE homolog (S. cerevisiae); K08516 s... 62.8 6e-10
pfa:PFI0515w PfYkt6.1; SNARE protein, putative; K08516 synapto... 61.2 2e-09
tgo:TGME49_099180 hypothetical protein ; K08516 synaptobrevin ... 60.1 4e-09
xla:735229 ykt6-b, ykt6b; YKT6 v-SNARE homolog (EC:2.3.1.-); K... 58.9 8e-09
dre:394067 MGC55536, Ykt6; zgc:55536 (EC:2.3.1.-); K08516 syna... 54.7 2e-07
tpv:TP02_0923 hypothetical protein; K08516 synaptobrevin homol... 49.7 5e-06
bbo:BBOV_I000460 16.m00764; hypothetical protein; K08516 synap... 45.4 9e-05
mmu:70358 Steap1, 2410007B19Rik, Prss24, Steap; six transmembr... 32.3 0.83
mmu:110095 Pygl; liver glycogen phosphorylase (EC:2.4.1.1); K0... 31.6 1.4
hsa:26872 STEAP1, MGC19484, PRSS24, STEAP; six transmembrane e... 31.6 1.5
eco:b1913 uvrC, ECK1912, JW1898; excinuclease UvrABC, endonucl... 31.6 1.6
hsa:5836 PYGL, GSD6; phosphorylase, glycogen, liver (EC:2.4.1.... 31.2 2.0
dre:553655 pygma, MGC110706, im:7150327, zgc:110706; phosphory... 30.8 2.5
dre:393444 pygmb, MGC63642, zgc:63642; phosphorylase, glycogen... 30.8 2.6
xla:494832 pygl; phosphorylase, glycogen, liver (EC:2.4.1.1); ... 30.4 3.1
xla:379862 pygm, MGC53328, pygb; phosphorylase, glycogen, musc... 30.4 3.7
dre:403051 pygb; phosphorylase, glycogen; brain (EC:2.4.1.1); ... 30.4 3.8
dre:493916 pygl, zgc:66314; phosphorylase, glycogen; liver (He... 30.4 3.8
xla:432134 hypothetical protein MGC80198; K00688 starch phosph... 30.0 3.9
mmu:110078 Pygb, MGC36329; brain glycogen phosphorylase (EC:2.... 29.6 5.0
hsa:5834 PYGB, MGC9213; phosphorylase, glycogen; brain (EC:2.4... 29.6 5.2
bbo:BBOV_III004950 17.m10540; hypothetical protein 29.3 6.9
mmu:223435 Trio, 6720464I07Rik, AA408740, Solo; triple functio... 28.9 9.4
> tgo:TGME49_015420 SNARE protein, putative ; K08516 synaptobrevin
homolog YKT6
Length=263
Score = 166 bits (420), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 87/186 (46%), Positives = 111/186 (59%), Gaps = 35/186 (18%)
Query 28 GAPEKSNIYALIIFKWDPEKPLLLSAAVDVSSFPFFHRSTMKEHIIFHSRLIAARTPPGR 87
GA K +Y+LI++KW PEKP+ L+ A D+SSFPFFHRSTMKEHI+FHSRLI ARTP GR
Sbjct 23 GANGKIALYSLILYKWSPEKPIQLATAFDLSSFPFFHRSTMKEHIVFHSRLICARTPLGR 82
Query 88 RQVVEFEKGLGRCFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQMQ----------- 136
RQVVEFE+ +G C VF H S L L++ YP+RVAFGL+ +AL+ Q
Sbjct 83 RQVVEFEQNIGHCHVFVHSSGLAATVLSTAAYPMRVAFGLITQALRGFQDRQGVENTCCG 142
Query 137 ----------------------QVEESAWRSLAADAKTDLL--PSSKLLLEKYKNPVEAD 172
Q+ W ++ D K ++ + LL+ Y+NPVEAD
Sbjct 143 TRSPRSTRHKCCEAHENPGDRWQIYAGQWENITDDVKEGIMFNKEASDLLKLYQNPVEAD 202
Query 173 KLLKVQ 178
KLLKVQ
Sbjct 203 KLLKVQ 208
> pfa:MAL13P1.135 PfYkt6.2; SNARE protein, putative; K08516 synaptobrevin
homolog YKT6
Length=221
Score = 127 bits (320), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 61/148 (41%), Positives = 100/148 (67%), Gaps = 4/148 (2%)
Query 35 IYALIIFKWDP-EKPLLLSAAVDVSSFPFFHRSTMKEHIIFHSRLIAARTPPGRRQVVEF 93
+Y+++I+K++ +P+ L++A+D+SSFPFFHRS++KEHI FH+RL+ RT G R+V+E
Sbjct 19 LYSILIYKYESSNQPIFLTSALDLSSFPFFHRSSLKEHIYFHARLVCGRTQKGTREVIEL 78
Query 94 EKGLGRCFVF-NHFSSLCCCALTSFNYPLRVAFGLLNEALKQMQQVEESAWRSLAADAKT 152
E G+G ++ N +++ L++ +YPLR+AF L++ K Q + + D K
Sbjct 79 ESGIGHLHIYTNKENNISVLVLSTSSYPLRIAFSLIDLTHKLFAQKCRGMYEHVRQDLKE 138
Query 153 DLLPSSKL--LLEKYKNPVEADKLLKVQ 178
+L ++L LL+KY+NP EADKL +VQ
Sbjct 139 GMLIQNELNDLLKKYQNPSEADKLSRVQ 166
> cpv:cgd6_510 synaptobrevin/VAMP-like protein ; K08516 synaptobrevin
homolog YKT6
Length=225
Score = 103 bits (257), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 51/166 (30%), Positives = 87/166 (52%), Gaps = 24/166 (14%)
Query 37 ALIIFKWDPEKPLLLSAAVDVSSFPFFHRSTMKEHIIFHSRLIAARTPPGRRQVVEFEKG 96
+++++KWD E P+LL++ ++ + FF R T++EHI FHSRL+ +R G R V F +
Sbjct 5 SVLLYKWDKEHPILLTSNYNLQDYNFFQRKTIQEHIAFHSRLLCSRVQQGNRVTVTFPQD 64
Query 97 LGRCFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQMQQVEESAWRSLAAD------- 149
+G C ++ + L C +TS +YP+R AF LLNE LK ++ ++D
Sbjct 65 IGHCHIYISNNGLGICCITSPDYPVRCAFSLLNEYLKTYIDSSKNHDVLFSSDFTTTTNN 124
Query 150 -----------------AKTDLLPSSKLLLEKYKNPVEADKLLKVQ 178
+ + +P + +KY++P+ DK+ KVQ
Sbjct 125 NNSNNNNNSIASNIITKDQNESIPECDEIFKKYQDPLNNDKIFKVQ 170
> ath:AT5G58060 YKT61; YKT61; K08516 synaptobrevin homolog YKT6
Length=221
Score = 100 bits (249), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 63/165 (38%), Positives = 88/165 (53%), Gaps = 22/165 (13%)
Query 35 IYALIIFKWDPE--KPLLLSAAVDVSSFPFFHRSTMKEHIIFHSRLIAARTPPGRRQVVE 92
I AL++ K PE P++LS A DVS F +F RS++KE ++F R +A+RTPP +RQ V+
Sbjct 3 ITALLVLKCAPEASDPVILSNASDVSHFGYFQRSSVKEFVVFVGRTVASRTPPSQRQSVQ 62
Query 93 FEK----------GLGRCFVFNHF---------SSLCCCALTSFNYPLRVAFGLLNEALK 133
E GL F F F + LC +YP+R AF LLN+ L
Sbjct 63 HEGCAPFLILDLPGLCPGFNFLRFYLIVHAYNRNGLCAVGFMDDHYPVRSAFSLLNQVLD 122
Query 134 QMQQVEESAWRSLAADAKTDLLPSSKLLLEKYKNPVEADKLLKVQ 178
+ Q+ +WRS D+ P L K+++P EADKLLK+Q
Sbjct 123 EYQKSFGESWRSAKEDSNQP-WPYLTEALNKFQDPAEADKLLKIQ 166
> ath:AT5G58180 ATYKT62; K08516 synaptobrevin homolog YKT6
Length=199
Score = 92.4 bits (228), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 56/148 (37%), Positives = 83/148 (56%), Gaps = 10/148 (6%)
Query 35 IYALIIFKWDPE--KPLLLSAAVDVSSFP--FFHRSTMKEHIIFHSRLIAARTPPGRRQV 90
I AL++ K DPE +P++L+ D+S F F+RS +E I+F +R +A RTPPG+RQ
Sbjct 3 ITALLVLKCDPETREPVILANVSDLSQFGKFSFYRSNFEEFIVFIARTVARRTPPGQRQS 62
Query 91 VEFEKGLGRCFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQMQQVEESAWRSLAADA 150
V+ E+ + N LC +YP+R AF LLN+ L Q+ WR +
Sbjct 63 VKHEEYKVHAYNIN---GLCAVGFMDDHYPVRSAFSLLNQVLDVYQKDYGDTWR---FEN 116
Query 151 KTDLLPSSKLLLEKYKNPVEADKLLKVQ 178
+ P K +K+++P EADKLLK+Q
Sbjct 117 SSQPWPYLKEASDKFRDPAEADKLLKIQ 144
> sce:YKL196C YKT6; Vesicle membrane protein (v-SNARE) with acyltransferase
activity; involved in trafficking to and within
the Golgi, endocytic trafficking to the vacuole, and vacuolar
fusion; membrane localization due to prenylation at the
carboxy-terminus (EC:2.3.1.-); K08516 synaptobrevin homolog
YKT6
Length=200
Score = 77.4 bits (189), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 44/145 (30%), Positives = 74/145 (51%), Gaps = 3/145 (2%)
Query 35 IYALIIFKWDPEKPLLLSAAVDVSSFPFFHRSTMKEHIIFHSRLIAARTPPGRRQVVEFE 94
IY + +F+ EK L LS D+S F FF RS++ + + F + +A+RT G+RQ +E
Sbjct 3 IYYIGVFRSGGEKALELSEVKDLSQFGFFERSSVGQFMTFFAETVASRTGAGQRQSIEEG 62
Query 95 KGLGRCFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQMQQVE-ESAWRSLAADAKTD 153
+G V+ +C +T YP+R A+ LLN+ L + + W +
Sbjct 63 NYIGH--VYARSEGICGVLITDKEYPVRPAYTLLNKILDEYLVAHPKEEWADVTETNDAL 120
Query 154 LLPSSKLLLEKYKNPVEADKLLKVQ 178
+ + KY++P +AD ++KVQ
Sbjct 121 KMKQLDTYISKYQDPSQADAIMKVQ 145
> xla:398034 ykt6-a, ykt6, ykt6a; YKT6 v-SNARE homolog (EC:2.3.1.-);
K08516 synaptobrevin homolog YKT6
Length=198
Score = 75.1 bits (183), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 53/146 (36%), Positives = 78/146 (53%), Gaps = 7/146 (4%)
Query 35 IYALIIFKWDPEKPLLLSAAVDVSSFPFFHRSTMKEHIIFHSRLIAARTPPGRRQVVEFE 94
+Y+L + K LL +A DVS F FF RS+++E + F S+LI R+ G R V+ +
Sbjct 3 LYSLSVLYKGENKVHLLKSAYDVSPFSFFQRSSIQEFMAFTSQLIVERSDKGSRSSVKEQ 62
Query 95 KGLGRCFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQMQ-QVEESAWRSLA-ADAKT 152
+ L C V+ SL + YP RV F LL + L++ QV+ W S + A +
Sbjct 63 EYL--CHVYVRNDSLAGVVIADNEYPPRVCFTLLEKVLEEFSTQVDRIDWPSGSPATIQY 120
Query 153 DLLPSSKLLLEKYKNPVEADKLLKVQ 178
+ L S L KY+NP +AD + KVQ
Sbjct 121 NALDS---YLSKYQNPRDADPMSKVQ 143
> cel:B0361.10 hypothetical protein; K08516 synaptobrevin homolog
YKT6
Length=201
Score = 68.9 bits (167), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 47/147 (31%), Positives = 74/147 (50%), Gaps = 6/147 (4%)
Query 35 IYALIIF--KWDPEKPLLLSAAVDVSSFPFFHRSTMKEHIIFHSRLIAARTPPGRRQVVE 92
+Y++++F D L + D+SSF FF R +++E + F ++L+ R+ G R V+
Sbjct 3 LYSILVFHKNVDTSDVKLFKSECDLSSFSFFQRGSVQEFMTFTAKLLVERSGLGARSSVK 62
Query 93 FEKGLGRCFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQ-MQQVEESAWRSLAADAK 151
+ L C+V N S C +T Y RVA L L +V + W + +D K
Sbjct 63 ENEYLVHCYVRNDGLSAVC--VTDAEYQQRVAMSFLGRVLDDFTTRVPATQWPGIRSD-K 119
Query 152 TDLLPSSKLLLEKYKNPVEADKLLKVQ 178
K LLEK++NP EAD + +VQ
Sbjct 120 DCSYTGLKDLLEKWQNPREADPMTRVQ 146
> mmu:56418 Ykt6, 0610042I15Rik, 1810013M05Rik, AW105923; YKT6
homolog (S. Cerevisiae); K08516 synaptobrevin homolog YKT6
Length=198
Score = 65.5 bits (158), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 60/149 (40%), Positives = 83/149 (55%), Gaps = 11/149 (7%)
Query 34 NIYAL-IIFKWDPEKPLLLSAAVDVSSFPFFHRSTMKEHIIFHSRLIAARTPPGRRQVVE 92
+Y+L +++K DP K +LL AA DVSSF FF RS+++E + F S+LI R+ G R V+
Sbjct 2 KLYSLSVLYKGDP-KAVLLKAAYDVSSFSFFQRSSVQEFMTFTSQLIVERSGKGSRASVK 60
Query 93 FEKGLGRCFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQM-QQVEESAW--RSLAAD 149
++ L C V+ SL + YP RVAF LL + L + +QV+ W S A
Sbjct 61 EQEYL--CHVYVRSDSLAGVVIADSEYPSRVAFTLLEKVLDEFSKQVDRIDWPVGSPATI 118
Query 150 AKTDLLPSSKLLLEKYKNPVEADKLLKVQ 178
T L L KY+NP EAD + KVQ
Sbjct 119 QYTGLDDH----LSKYQNPREADPMSKVQ 143
> hsa:10652 YKT6; YKT6 v-SNARE homolog (S. cerevisiae); K08516
synaptobrevin homolog YKT6
Length=198
Score = 62.8 bits (151), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 54/145 (37%), Positives = 76/145 (52%), Gaps = 5/145 (3%)
Query 35 IYALIIFKWDPEKPLLLSAAVDVSSFPFFHRSTMKEHIIFHSRLIAARTPPGRRQVVEFE 94
+Y+L + K +LL AA DVSSF FF RS+++E + F S+LI R+ G R V+ +
Sbjct 3 LYSLSVLYKGEAKVVLLKAAYDVSSFSFFQRSSVQEFMTFTSQLIVERSSKGTRASVKEQ 62
Query 95 KGLGRCFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQM-QQVEESAWRSLAADAKTD 153
L C V+ SL + YP RVAF LL + L + +QV+ W T
Sbjct 63 DYL--CHVYVRNDSLAGVVIADNEYPSRVAFTLLEKVLDEFSKQVDRIDWP--VGSPATI 118
Query 154 LLPSSKLLLEKYKNPVEADKLLKVQ 178
P+ L +Y+NP EAD + KVQ
Sbjct 119 HYPALDGHLSRYQNPREADPMTKVQ 143
> pfa:PFI0515w PfYkt6.1; SNARE protein, putative; K08516 synaptobrevin
homolog YKT6
Length=199
Score = 61.2 bits (147), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 37/146 (25%), Positives = 70/146 (47%), Gaps = 6/146 (4%)
Query 34 NIYALIIFKWDPEKPLLLSAAVDVSSFPFFHRSTMKEHIIFHSRLIAARTPPGRRQVVEF 93
N+ A+ + K+ E + L A D++S+ F + KE F +R I +R ++++
Sbjct 2 NLLAIFLTKY-VEDVIFLCNATDLNSYSFIKKKAFKEAAFFVARTIPSRIEYNTKEIITH 60
Query 94 EKGLGRCFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQ-MQQVEESAWRSLAADAKT 152
E F F + ++C + + +YP RVAF ++NE + + + + W S+ D K
Sbjct 61 ENN--TVFAFKYEDNICPIVIATDDYPERVAFYMINEIYRDFISTIPKEEWSSVKQDNKI 118
Query 153 DLLPSSKLLLEKYKNPVEADKLLKVQ 178
+ L KYK+P+ D + +
Sbjct 119 SF--NLYTYLTKYKDPLNCDAITQTN 142
> tgo:TGME49_099180 hypothetical protein ; K08516 synaptobrevin
homolog YKT6
Length=211
Score = 60.1 bits (144), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 42/141 (29%), Positives = 69/141 (48%), Gaps = 9/141 (6%)
Query 44 DPEKPLLLSAAVDVSSFPFFHRSTMKEHIIFHSRLIAARTPPGRRQVVEFEKGLGRCFVF 103
D + P+ L +A ++ + F+ R +MKE F +R R R+V+E + + F++
Sbjct 14 DADAPVFLCSAFELGGYAFYQRHSMKEACKFLARTAIPRVGVETREVIEHDGSVA--FLY 71
Query 104 NHFSSLCCCALTSFNYPLRVAFGLLNEALKQMQQ-VEESAWRSLA------ADAKTDLLP 156
F SL A+ NYP RVAF LL+E ++ + V + W S+A A+
Sbjct 72 RFFDSLAVVAIGDANYPSRVAFRLLHEIHEKFTRAVPVALWSSVAPVGNSSCAAQIHFKD 131
Query 157 SSKLLLEKYKNPVEADKLLKV 177
LLL + ++ +AD L V
Sbjct 132 DLALLLRRCQDARKADTLTAV 152
> xla:735229 ykt6-b, ykt6b; YKT6 v-SNARE homolog (EC:2.3.1.-);
K08516 synaptobrevin homolog YKT6
Length=198
Score = 58.9 bits (141), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 54/146 (36%), Positives = 79/146 (54%), Gaps = 7/146 (4%)
Query 35 IYALIIFKWDPEKPLLLSAAVDVSSFPFFHRSTMKEHIIFHSRLIAARTPPGRRQVVEFE 94
+Y+L + K LL +A DVSSF FF RS+++E + F S+LI R+ G R V+ +
Sbjct 3 LYSLSVLYKGENKVHLLKSAYDVSSFSFFQRSSVQEFMTFTSQLIVERSDKGSRSSVKEQ 62
Query 95 KGLGRCFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQMQ-QVEESAWRSLA-ADAKT 152
+ L C V+ SL + YP RV F LL + L++ QV+ W S + A +
Sbjct 63 EYL--CHVYVRNDSLAGVVIADNEYPPRVCFTLLEKVLEEFSTQVDRIDWPSGSPATIQY 120
Query 153 DLLPSSKLLLEKYKNPVEADKLLKVQ 178
+ L S L KY+NP +AD + KVQ
Sbjct 121 NALDS---YLSKYQNPRDADPMSKVQ 143
> dre:394067 MGC55536, Ykt6; zgc:55536 (EC:2.3.1.-); K08516 synaptobrevin
homolog YKT6
Length=198
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 52/146 (35%), Positives = 76/146 (52%), Gaps = 7/146 (4%)
Query 35 IYALIIFKWDPEKPLLLSAAVDVSSFPFFHRSTMKEHIIFHSRLIAARTPPGRRQVVEFE 94
+Y+L + K LL A D+SSF FF RS+++E + F S LI R+ G R V+ +
Sbjct 3 LYSLSVLHKGSTKANLLKATYDLSSFSFFQRSSVQEFMTFTSALIVERSALGSRASVKEQ 62
Query 95 KGLGRCFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQM-QQVEESAWRSLA-ADAKT 152
+ L C V+ +L + YP RV F LL++ L + +QV W S + A +
Sbjct 63 EYL--CHVYVRNDNLGGVVIADSEYPSRVCFTLLDKVLDEFSRQVNSIDWPSGSPATIQY 120
Query 153 DLLPSSKLLLEKYKNPVEADKLLKVQ 178
L S L +Y+NP EAD + KVQ
Sbjct 121 TALDSH---LARYQNPREADAMTKVQ 143
> tpv:TP02_0923 hypothetical protein; K08516 synaptobrevin homolog
YKT6
Length=201
Score = 49.7 bits (117), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 44/145 (30%), Positives = 65/145 (44%), Gaps = 11/145 (7%)
Query 31 EKSNIYALIIFKWDPEKPLLLSAAVDVSSFPFFHRSTMKEHIIFHSRLIAARTPPGRRQV 90
+K NI L + + E+P+ L+ S P+F R K F +R ++ R G
Sbjct 2 QKHNITYLGVLQC-KEEPVFLTQCFHFSDLPYFSRGQAKNVSTFMARELSKRVDYGFTAT 60
Query 91 VEFEKGLGRCFVF-NHFSS-LCCCALTSFNYPLRVAFGLLNEALKQMQQVEESAWRSLAA 148
+ V+ N +SS LC + + +YP R AFGLL + E+ + L
Sbjct 61 T-----VDDYLVYSNKWSSGLCILCICNRDYPSRTAFGLLQHSFFLFN--EKYVYEELKF 113
Query 149 DAKTDL-LPSSKLLLEKYKNPVEAD 172
D +L LP K L+ KYKNP D
Sbjct 114 DNDLNLSLPELKELMLKYKNPEAVD 138
> bbo:BBOV_I000460 16.m00764; hypothetical protein; K08516 synaptobrevin
homolog YKT6
Length=202
Score = 45.4 bits (106), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 36/132 (27%), Positives = 52/132 (39%), Gaps = 3/132 (2%)
Query 46 EKPLLLSAAVDVSSFPFFHRSTMKEHIIFHSRLIAARTPPGRRQVVEFEKGLGRCFVFNH 105
EKP+ L +S R T+K +F +R IA R G V E F +
Sbjct 16 EKPIFLLQYFKLSDLQILSRGTVKSVAVFTAREIAGRLNQGENVAVHEEN--FDVFAYRW 73
Query 106 FSSLCCCALTSFNYPLRVAFGLLNEALKQMQQVEESAWRSLAADAKTDLLPSSKLLLEKY 165
+C + YP RVAF LL A + A +D + +P K L+ +Y
Sbjct 74 DVGVCAICICGKGYPERVAFSLLQLAFFEFITKYPDAGIEHTSDVNLN-IPQIKALVNQY 132
Query 166 KNPVEADKLLKV 177
++P D V
Sbjct 133 RDPTVVDAYTNV 144
> mmu:70358 Steap1, 2410007B19Rik, Prss24, Steap; six transmembrane
epithelial antigen of the prostate 1; K14737 metalloreductase
STEAP1 [EC:1.16.1.-]
Length=339
Score = 32.3 bits (72), Expect = 0.83, Method: Compositional matrix adjust.
Identities = 16/33 (48%), Positives = 23/33 (69%), Gaps = 2/33 (6%)
Query 113 ALTSFNYPLRVAF--GLLNEALKQMQQVEESAW 143
A+ S +YP+R ++ LLN A KQ+QQ +E AW
Sbjct 176 AVYSLSYPMRRSYRYKLLNWAYKQVQQNKEDAW 208
> mmu:110095 Pygl; liver glycogen phosphorylase (EC:2.4.1.1);
K00688 starch phosphorylase [EC:2.4.1.1]
Length=850
Score = 31.6 bits (70), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 12/42 (28%), Positives = 24/42 (57%), Gaps = 4/42 (9%)
Query 100 CFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQMQQVEES 141
CF+ S+ L ++ Y +R +G+ N+ +++ QVEE+
Sbjct 143 CFL----DSMATLGLAAYGYGIRYEYGIFNQKIREGWQVEEA 180
> hsa:26872 STEAP1, MGC19484, PRSS24, STEAP; six transmembrane
epithelial antigen of the prostate 1; K14737 metalloreductase
STEAP1 [EC:1.16.1.-]
Length=339
Score = 31.6 bits (70), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 23/33 (69%), Gaps = 2/33 (6%)
Query 113 ALTSFNYPLRVAF--GLLNEALKQMQQVEESAW 143
A+ S +YP+R ++ LLN A +Q+QQ +E AW
Sbjct 176 AIYSLSYPMRRSYRYKLLNWAYQQVQQNKEDAW 208
> eco:b1913 uvrC, ECK1912, JW1898; excinuclease UvrABC, endonuclease
subunit; K03703 excinuclease ABC subunit C
Length=610
Score = 31.6 bits (70), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 28/60 (46%), Gaps = 6/60 (10%)
Query 1 DSAKLNSFFNLKTPE-MGDSGVPA-----GGEGGAPEKSNIYALIIFKWDPEKPLLLSAA 54
D A +N + + + DS +P GG+G + N++A + WD PLLL A
Sbjct 438 DYAAMNQVLRRRYGKAIDDSKIPDVILIDGGKGQLAQAKNVFAELDVSWDKNHPLLLGVA 497
> hsa:5836 PYGL, GSD6; phosphorylase, glycogen, liver (EC:2.4.1.1);
K00688 starch phosphorylase [EC:2.4.1.1]
Length=813
Score = 31.2 bits (69), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 12/42 (28%), Positives = 23/42 (54%), Gaps = 4/42 (9%)
Query 100 CFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQMQQVEES 141
CF+ S+ L ++ Y +R +G+ N+ ++ QVEE+
Sbjct 109 CFL----DSMATLGLAAYGYGIRYEYGIFNQKIRDGWQVEEA 146
> dre:553655 pygma, MGC110706, im:7150327, zgc:110706; phosphorylase,
glycogen (muscle) A (EC:2.4.1.1); K00688 starch phosphorylase
[EC:2.4.1.1]
Length=842
Score = 30.8 bits (68), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 13/42 (30%), Positives = 22/42 (52%), Gaps = 4/42 (9%)
Query 100 CFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQMQQVEES 141
CF+ S+ L ++ Y +R FG+ N+ + QVEE+
Sbjct 143 CFL----DSMASLGLAAYGYGIRYEFGIFNQKISNGWQVEEA 180
> dre:393444 pygmb, MGC63642, zgc:63642; phosphorylase, glycogen
(muscle) b
Length=315
Score = 30.8 bits (68), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 13/42 (30%), Positives = 23/42 (54%), Gaps = 4/42 (9%)
Query 100 CFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQMQQVEES 141
CF+ S+ L ++ Y +R FG+ N+ + + QVEE+
Sbjct 143 CFL----DSMATLGLAAYGYGIRYEFGIFNQKIVKGWQVEEA 180
> xla:494832 pygl; phosphorylase, glycogen, liver (EC:2.4.1.1);
K00688 starch phosphorylase [EC:2.4.1.1]
Length=855
Score = 30.4 bits (67), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 12/42 (28%), Positives = 22/42 (52%), Gaps = 4/42 (9%)
Query 100 CFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQMQQVEES 141
CF+ S+ L ++ Y +R +G+ N+ +K Q EE+
Sbjct 143 CFL----DSMATLGLAAYGYGIRYEYGIFNQKIKDGWQAEEA 180
> xla:379862 pygm, MGC53328, pygb; phosphorylase, glycogen, muscle
(EC:2.4.1.1); K00688 starch phosphorylase [EC:2.4.1.1]
Length=843
Score = 30.4 bits (67), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 13/42 (30%), Positives = 22/42 (52%), Gaps = 4/42 (9%)
Query 100 CFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQMQQVEES 141
CF+ S+ L ++ Y +R FG+ N+ + QVEE+
Sbjct 143 CFL----DSMATLGLAAYGYGIRYEFGIFNQRIMNGWQVEEA 180
> dre:403051 pygb; phosphorylase, glycogen; brain (EC:2.4.1.1);
K00688 starch phosphorylase [EC:2.4.1.1]
Length=843
Score = 30.4 bits (67), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 12/42 (28%), Positives = 22/42 (52%), Gaps = 4/42 (9%)
Query 100 CFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQMQQVEES 141
CF+ S+ L ++ Y +R FG+ N+ + Q+EE+
Sbjct 143 CFL----DSMASLGLAAYGYGIRYEFGIFNQKIAHGWQIEEA 180
> dre:493916 pygl, zgc:66314; phosphorylase, glycogen; liver (Hers
disease, glycogen storage disease type VI) (EC:2.4.1.1);
K00688 starch phosphorylase [EC:2.4.1.1]
Length=967
Score = 30.4 bits (67), Expect = 3.8, Method: Composition-based stats.
Identities = 18/69 (26%), Positives = 32/69 (46%), Gaps = 15/69 (21%)
Query 88 RQVVEFEKGLGR-------CFVFNH--------FSSLCCCALTSFNYPLRVAFGLLNEAL 132
R++ E+G G+ C V +H S+ L ++ Y +R +G+ N+ +
Sbjct 426 RRMSPIEEGGGKRVNMAHLCIVGSHKVNGAACFLDSMATLGLAAYGYGIRYEYGIFNQKI 485
Query 133 KQMQQVEES 141
K QVEE+
Sbjct 486 KDGWQVEEA 494
Score = 29.6 bits (65), Expect = 5.6, Method: Composition-based stats.
Identities = 13/42 (30%), Positives = 23/42 (54%), Gaps = 4/42 (9%)
Query 100 CFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQMQQVEES 141
CF+ S+ L ++ Y +R +G+ N+ +K QVEE+
Sbjct 143 CFL----DSMATLGLAAYGYGIRYEYGIFNQKIKDGWQVEEA 180
> xla:432134 hypothetical protein MGC80198; K00688 starch phosphorylase
[EC:2.4.1.1]
Length=843
Score = 30.0 bits (66), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 13/42 (30%), Positives = 22/42 (52%), Gaps = 4/42 (9%)
Query 100 CFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQMQQVEES 141
CF+ S+ L ++ Y +R FG+ N+ + QVEE+
Sbjct 143 CFL----DSMATLGLAAYGYGIRYEFGIFNQRIMNGWQVEEA 180
> mmu:110078 Pygb, MGC36329; brain glycogen phosphorylase (EC:2.4.1.1);
K00688 starch phosphorylase [EC:2.4.1.1]
Length=843
Score = 29.6 bits (65), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 13/42 (30%), Positives = 22/42 (52%), Gaps = 4/42 (9%)
Query 100 CFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQMQQVEES 141
CF+ S+ L ++ Y +R FG+ N+ + QVEE+
Sbjct 143 CFL----DSMATLGLAAYGYGIRYEFGIFNQKIVNGWQVEEA 180
> hsa:5834 PYGB, MGC9213; phosphorylase, glycogen; brain (EC:2.4.1.1);
K00688 starch phosphorylase [EC:2.4.1.1]
Length=843
Score = 29.6 bits (65), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 13/42 (30%), Positives = 22/42 (52%), Gaps = 4/42 (9%)
Query 100 CFVFNHFSSLCCCALTSFNYPLRVAFGLLNEALKQMQQVEES 141
CF+ S+ L ++ Y +R FG+ N+ + QVEE+
Sbjct 143 CFL----DSMATLGLAAYGYGIRYEFGIFNQKIVNGWQVEEA 180
> bbo:BBOV_III004950 17.m10540; hypothetical protein
Length=880
Score = 29.3 bits (64), Expect = 6.9, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 21/37 (56%), Gaps = 0/37 (0%)
Query 26 EGGAPEKSNIYALIIFKWDPEKPLLLSAAVDVSSFPF 62
E PE+ ALI + PEKPL+ A +D+++ +
Sbjct 175 ESKIPERYGPVALIAIRVIPEKPLIQDALIDINNLVY 211
> mmu:223435 Trio, 6720464I07Rik, AA408740, Solo; triple functional
domain (PTPRF interacting) (EC:2.7.11.1); K08810 triple
functional domain protein [EC:2.7.11.1]
Length=3103
Score = 28.9 bits (63), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 38/79 (48%), Gaps = 11/79 (13%)
Query 104 NHFSSLCCCALTSFNYPLRVAFGLL------NEALKQMQQVEESAWRSLAA--DAKTDLL 155
NHF+ C + N + VA L+ ++ +KQ+ E W++ AA D ++ LL
Sbjct 385 NHFAMNCMNVYVNINRIMSVANRLVESGHYASQQIKQIANQLEQEWKAFAAALDERSTLL 444
Query 156 PSSKLL---LEKYKNPVEA 171
S + EKY + V++
Sbjct 445 DMSSIFHQKAEKYMSNVDS 463
Lambda K H
0.320 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4730349484
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40