bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_7940_orf1
Length=59
Score E
Sequences producing significant alignments: (Bits) Value
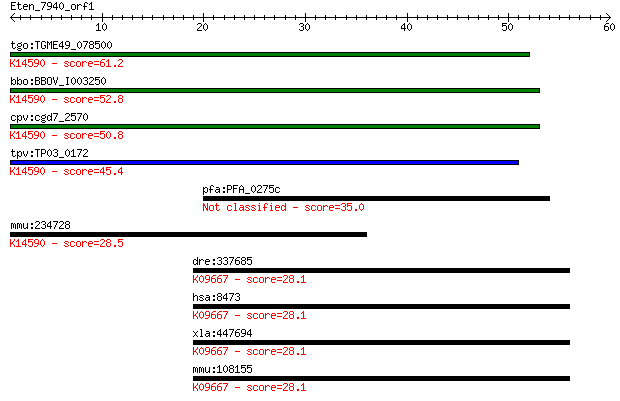
tgo:TGME49_078500 ftsJ-like methyltransferase domain-containin... 61.2 8e-10
bbo:BBOV_I003250 19.m02092; ribosomal RNA large subunit methyl... 52.8 2e-07
cpv:cgd7_2570 adrift-like. FTSJ family RNA methylase ; K14590 ... 50.8 1e-06
tpv:TP03_0172 hypothetical protein; K14590 FtsJ methyltransfer... 45.4 5e-05
pfa:PFA_0275c conserved Plasmodium protein, unknown function 35.0 0.059
mmu:234728 Ftsjd1, AU022703, C730036L12Rik, MGC38131; FtsJ met... 28.5 6.0
dre:337685 ogt.1, fm81g08, ogt, wu:fc12b01, wu:fm81g08; O-link... 28.1 6.5
hsa:8473 OGT, FLJ23071, HRNT1, MGC22921, O-GLCNAC; O-linked N-... 28.1 6.7
xla:447694 ogt, MGC80426; O-linked N-acetylglucosamine (GlcNAc... 28.1 7.0
mmu:108155 Ogt, 1110038P24Rik, 4831420N21Rik, AI115525, Ogtl; ... 28.1 7.1
> tgo:TGME49_078500 ftsJ-like methyltransferase domain-containing
protein ; K14590 FtsJ methyltransferase domain-containing
protein 1 [EC:2.1.1.-]
Length=1422
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 26/51 (50%), Positives = 36/51 (70%), Gaps = 0/51 (0%)
Query 1 PYFEGCSPLEVLDDDELYRHSERRWISSPDGSGNICKKENIEFLWKQLTRQ 51
PY EG +P E+LDDD LYR +E W++ D +G+I KK NIEF+W +R+
Sbjct 227 PYHEGGNPSELLDDDVLYRQTEDLWLTGYDETGDIVKKRNIEFIWNHTSRE 277
> bbo:BBOV_I003250 19.m02092; ribosomal RNA large subunit methyltransferase
J domain containing protein; K14590 FtsJ methyltransferase
domain-containing protein 1 [EC:2.1.1.-]
Length=1005
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 23/52 (44%), Positives = 34/52 (65%), Gaps = 0/52 (0%)
Query 1 PYFEGCSPLEVLDDDELYRHSERRWISSPDGSGNICKKENIEFLWKQLTRQA 52
PY EG S +E L +D LYR + W++ D SGNI K NIE++W +++R +
Sbjct 197 PYHEGNSHMECLAEDILYRDTVTNWLTGSDESGNINKTANIEYIWDRVSRAS 248
> cpv:cgd7_2570 adrift-like. FTSJ family RNA methylase ; K14590
FtsJ methyltransferase domain-containing protein 1 [EC:2.1.1.-]
Length=1188
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 33/52 (63%), Gaps = 0/52 (0%)
Query 1 PYFEGCSPLEVLDDDELYRHSERRWISSPDGSGNICKKENIEFLWKQLTRQA 52
PYFEG +P +L DD +YR + R W D SG+I K NI+++W++ ++
Sbjct 177 PYFEGNNPNAILTDDIIYRDTYRAWNMGADDSGDITKVCNIDYIWQKTSKNT 228
> tpv:TP03_0172 hypothetical protein; K14590 FtsJ methyltransferase
domain-containing protein 1 [EC:2.1.1.-]
Length=980
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 20/50 (40%), Positives = 31/50 (62%), Gaps = 0/50 (0%)
Query 1 PYFEGCSPLEVLDDDELYRHSERRWISSPDGSGNICKKENIEFLWKQLTR 50
PY+EG + VL +D L + + + W+ D SGNI K NIE++W ++R
Sbjct 196 PYYEGNNHNVVLAEDILLKETYQNWLLGYDNSGNITKSCNIEYIWDHISR 245
> pfa:PFA_0275c conserved Plasmodium protein, unknown function
Length=1224
Score = 35.0 bits (79), Expect = 0.059, Method: Composition-based stats.
Identities = 12/34 (35%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 20 HSERRWISSPDGSGNICKKENIEFLWKQLTRQAH 53
+ +WIS + +GNI ENIE++W + R+ +
Sbjct 248 YENEKWISGINNTGNILHLENIEYVWNKTVREGN 281
> mmu:234728 Ftsjd1, AU022703, C730036L12Rik, MGC38131; FtsJ methyltransferase
domain containing 1; K14590 FtsJ methyltransferase
domain-containing protein 1 [EC:2.1.1.-]
Length=767
Score = 28.5 bits (62), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 18/35 (51%), Gaps = 0/35 (0%)
Query 1 PYFEGCSPLEVLDDDELYRHSERRWISSPDGSGNI 35
PY E L ++ DD L ++ W PD +G+I
Sbjct 176 PYHEANDNLRMITDDRLMANTLHCWYFGPDNTGDI 210
> dre:337685 ogt.1, fm81g08, ogt, wu:fc12b01, wu:fm81g08; O-linked
N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl
transferase) 1;
K09667 polypeptide N-acetylglucosaminyltransferase [EC:2.4.1.-]
Length=1062
Score = 28.1 bits (61), Expect = 6.5, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 19/37 (51%), Gaps = 0/37 (0%)
Query 19 RHSERRWISSPDGSGNICKKENIEFLWKQLTRQAHPS 55
RH + W PD +G + +I F ++L R AH S
Sbjct 42 RHCMQLWRQEPDNTGVLLLLSSIHFQCRRLDRSAHFS 78
> hsa:8473 OGT, FLJ23071, HRNT1, MGC22921, O-GLCNAC; O-linked
N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl
transferase); K09667
polypeptide N-acetylglucosaminyltransferase [EC:2.4.1.-]
Length=1046
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 19/37 (51%), Gaps = 0/37 (0%)
Query 19 RHSERRWISSPDGSGNICKKENIEFLWKQLTRQAHPS 55
RH + W PD +G + +I F ++L R AH S
Sbjct 42 RHCMQLWRQEPDNTGVLLLLSSIHFQCRRLDRSAHFS 78
> xla:447694 ogt, MGC80426; O-linked N-acetylglucosamine (GlcNAc)
transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl
transferase); K09667 polypeptide N-acetylglucosaminyltransferase
[EC:2.4.1.-]
Length=1063
Score = 28.1 bits (61), Expect = 7.0, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 19/37 (51%), Gaps = 0/37 (0%)
Query 19 RHSERRWISSPDGSGNICKKENIEFLWKQLTRQAHPS 55
RH + W PD +G + +I F ++L R AH S
Sbjct 42 RHCMQLWRQEPDNTGVLLLLSSIHFQCRRLDRSAHFS 78
> mmu:108155 Ogt, 1110038P24Rik, 4831420N21Rik, AI115525, Ogtl;
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl
transferase);
K09667 polypeptide N-acetylglucosaminyltransferase [EC:2.4.1.-]
Length=1046
Score = 28.1 bits (61), Expect = 7.1, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 19/37 (51%), Gaps = 0/37 (0%)
Query 19 RHSERRWISSPDGSGNICKKENIEFLWKQLTRQAHPS 55
RH + W PD +G + +I F ++L R AH S
Sbjct 42 RHCMQLWRQEPDNTGVLLLLSSIHFQCRRLDRSAHFS 78
Lambda K H
0.318 0.139 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2073802932
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40