bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8010_orf1
Length=70
Score E
Sequences producing significant alignments: (Bits) Value
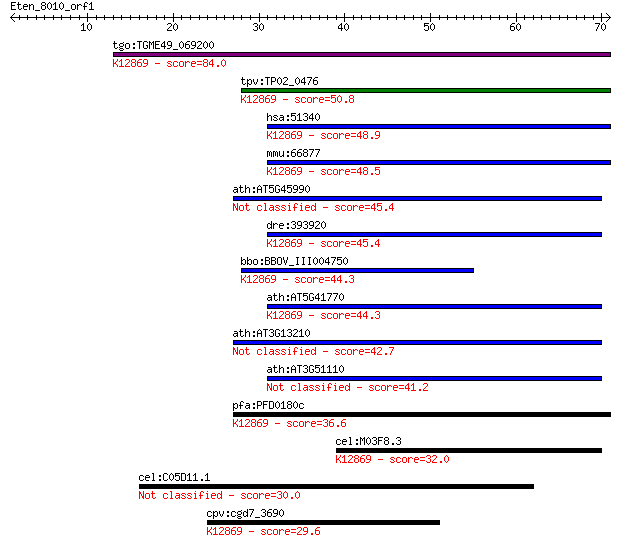
tgo:TGME49_069200 crooked neck-like protein 1, putative ; K128... 84.0 1e-16
tpv:TP02_0476 crooked neck protein; K12869 crooked neck 50.8 1e-06
hsa:51340 CRNKL1, CLF, CRN, Clf1, HCRN, MSTP021, SYF3; crooked... 48.9 4e-06
mmu:66877 Crnkl1, 1200013P10Rik, 5730590A01Rik, C80326, crn; C... 48.5 5e-06
ath:AT5G45990 crooked neck protein, putative / cell cycle prot... 45.4 4e-05
dre:393920 crnkl1, MGC55327, zgc:55327; crooked neck pre-mRNA ... 45.4 4e-05
bbo:BBOV_III004750 17.m07426; tetratricopeptide repeat domain ... 44.3 8e-05
ath:AT5G41770 crooked neck protein, putative / cell cycle prot... 44.3 1e-04
ath:AT3G13210 crooked neck protein, putative / cell cycle prot... 42.7 3e-04
ath:AT3G51110 crooked neck protein, putative / cell cycle prot... 41.2 8e-04
pfa:PFD0180c CGI-201 protein, short form; K12869 crooked neck 36.6 0.020
cel:M03F8.3 hypothetical protein; K12869 crooked neck 32.0 0.45
cel:C05D11.1 hypothetical protein 30.0 2.0
cpv:cgd7_3690 crooked neck protein HAT repeats ; K12869 crooke... 29.6 2.5
> tgo:TGME49_069200 crooked neck-like protein 1, putative ; K12869
crooked neck
Length=794
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 42/58 (72%), Positives = 45/58 (77%), Gaps = 0/58 (0%)
Query 13 PRRGAKNYPLGASWKQMEVKNKMPAAVQITAEQLLREAVDRQLEDAAAAKPQQRIVDE 70
P NYPL S K MEVKNKMPA VQITAEQLLREAVDRQL+D + +PQQRIVDE
Sbjct 15 PESAGGNYPLHRSRKMMEVKNKMPAPVQITAEQLLREAVDRQLDDLSQIRPQQRIVDE 72
> tpv:TP02_0476 crooked neck protein; K12869 crooked neck
Length=657
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 25/43 (58%), Positives = 33/43 (76%), Gaps = 2/43 (4%)
Query 28 QMEVKNKMPAAVQITAEQLLREAVDRQLEDAAAAKPQQRIVDE 70
+++VKNKMPAAVQITAEQ+LR+AV+ Q ++ K Q I DE
Sbjct 8 KLQVKNKMPAAVQITAEQILRDAVEWQTKEVKTTK--QTIADE 48
> hsa:51340 CRNKL1, CLF, CRN, Clf1, HCRN, MSTP021, SYF3; crooked
neck pre-mRNA splicing factor-like 1 (Drosophila); K12869
crooked neck
Length=848
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 27/40 (67%), Positives = 30/40 (75%), Gaps = 1/40 (2%)
Query 31 VKNKMPAAVQITAEQLLREAVDRQLEDAAAAKPQQRIVDE 70
VKNK PA VQITAEQLLREA +R+LE PQQ+I DE
Sbjct 179 VKNKAPAEVQITAEQLLREAKERELE-LLPPPPQQKITDE 217
> mmu:66877 Crnkl1, 1200013P10Rik, 5730590A01Rik, C80326, crn;
Crn, crooked neck-like 1 (Drosophila); K12869 crooked neck
Length=690
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 27/40 (67%), Positives = 30/40 (75%), Gaps = 1/40 (2%)
Query 31 VKNKMPAAVQITAEQLLREAVDRQLEDAAAAKPQQRIVDE 70
VKNK PA VQITAEQLLREA +R+LE PQQ+I DE
Sbjct 18 VKNKAPAEVQITAEQLLREAKERELE-LLPPPPQQKITDE 56
> ath:AT5G45990 crooked neck protein, putative / cell cycle protein,
putative
Length=673
Score = 45.4 bits (106), Expect = 4e-05, Method: Composition-based stats.
Identities = 24/43 (55%), Positives = 30/43 (69%), Gaps = 2/43 (4%)
Query 27 KQMEVKNKMPAAVQITAEQLLREAVDRQLEDAAAAKPQQRIVD 69
+ VKNK PA VQITAEQ+LREA +RQ +A P+Q+I D
Sbjct 12 RTTRVKNKTPAPVQITAEQILREARERQ--EAEIRPPKQKITD 52
> dre:393920 crnkl1, MGC55327, zgc:55327; crooked neck pre-mRNA
splicing factor-like 1 (Drosophila); K12869 crooked neck
Length=753
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 25/39 (64%), Positives = 29/39 (74%), Gaps = 1/39 (2%)
Query 31 VKNKMPAAVQITAEQLLREAVDRQLEDAAAAKPQQRIVD 69
VKNK PA VQITAEQLLREA +R+LE P+Q+I D
Sbjct 17 VKNKAPAEVQITAEQLLREAKERELE-LLPPPPKQKITD 54
> bbo:BBOV_III004750 17.m07426; tetratricopeptide repeat domain
containing protein; K12869 crooked neck
Length=665
Score = 44.3 bits (103), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 20/27 (74%), Positives = 26/27 (96%), Gaps = 0/27 (0%)
Query 28 QMEVKNKMPAAVQITAEQLLREAVDRQ 54
+++VKNKMPAAVQITAEQ+LR+AV+ Q
Sbjct 8 KLQVKNKMPAAVQITAEQILRDAVEWQ 34
> ath:AT5G41770 crooked neck protein, putative / cell cycle protein,
putative; K12869 crooked neck
Length=705
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 23/39 (58%), Positives = 29/39 (74%), Gaps = 2/39 (5%)
Query 31 VKNKMPAAVQITAEQLLREAVDRQLEDAAAAKPQQRIVD 69
VKNK PA +QITAEQ+LREA +RQ +A P+Q+I D
Sbjct 30 VKNKTPAPIQITAEQILREARERQ--EAEIRPPKQKITD 66
> ath:AT3G13210 crooked neck protein, putative / cell cycle protein,
putative
Length=657
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 23/43 (53%), Positives = 29/43 (67%), Gaps = 2/43 (4%)
Query 27 KQMEVKNKMPAAVQITAEQLLREAVDRQLEDAAAAKPQQRIVD 69
+ +VKNK PA +QITAEQ+LREA +RQ +A P Q I D
Sbjct 25 RMTQVKNKTPAPIQITAEQILREARERQ--EAEFRPPNQTITD 65
> ath:AT3G51110 crooked neck protein, putative / cell cycle protein,
putative
Length=413
Score = 41.2 bits (95), Expect = 8e-04, Method: Composition-based stats.
Identities = 23/40 (57%), Positives = 30/40 (75%), Gaps = 3/40 (7%)
Query 31 VKNKMPAAVQITAEQLLREAVDRQLEDAAAAK-PQQRIVD 69
VKNK PA VQITAEQ+L+EA R+ ED+ + P+Q+I D
Sbjct 8 VKNKTPAPVQITAEQVLKEA--REREDSRILRPPKQKITD 45
> pfa:PFD0180c CGI-201 protein, short form; K12869 crooked neck
Length=780
Score = 36.6 bits (83), Expect = 0.020, Method: Composition-based stats.
Identities = 21/44 (47%), Positives = 30/44 (68%), Gaps = 3/44 (6%)
Query 27 KQMEVKNKMPAAVQITAEQLLREAVDRQLEDAAAAKPQQRIVDE 70
K+++VKNK A VQITAEQL+ EA+ +LE+ K ++DE
Sbjct 6 KKLQVKNKNAAEVQITAEQLINEAL--ELEE-VEHKVNYNLIDE 46
> cel:M03F8.3 hypothetical protein; K12869 crooked neck
Length=747
Score = 32.0 bits (71), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 18/31 (58%), Positives = 23/31 (74%), Gaps = 1/31 (3%)
Query 39 VQITAEQLLREAVDRQLEDAAAAKPQQRIVD 69
+QITAEQLLREA +R+LE A P+ +I D
Sbjct 31 LQITAEQLLREAKERELELIPPA-PKTKITD 60
> cel:C05D11.1 hypothetical protein
Length=995
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 25/48 (52%), Gaps = 2/48 (4%)
Query 16 GAKNYPLGASWKQMEVKNKM--PAAVQITAEQLLREAVDRQLEDAAAA 61
GA YPL A W Q+ + + P+ + A++L EA DR+ + A
Sbjct 633 GADKYPLLAKWAQIFTQGVVFDPSRIHQCAQKLAGEARDRKRDGCTVA 680
> cpv:cgd7_3690 crooked neck protein HAT repeats ; K12869 crooked
neck
Length=736
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 13/27 (48%), Positives = 20/27 (74%), Gaps = 0/27 (0%)
Query 24 ASWKQMEVKNKMPAAVQITAEQLLREA 50
A+ ++ VKNK A +QIT EQ+L+E+
Sbjct 25 ANEQESNVKNKSFAPIQITVEQILKES 51
Lambda K H
0.313 0.130 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2024947620
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40