bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8027_orf1
Length=103
Score E
Sequences producing significant alignments: (Bits) Value
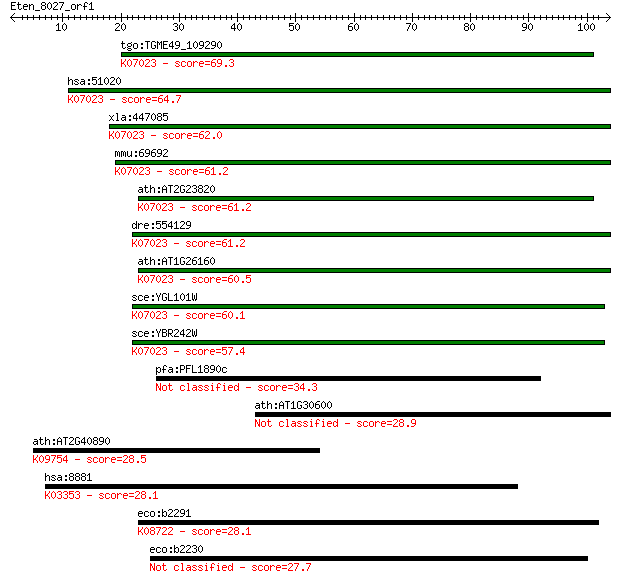
tgo:TGME49_109290 HD domain-containing protein ; K07023 putati... 69.3 3e-12
hsa:51020 HDDC2, C6orf74, MGC87330, NS5ATP2, dJ167O5.2; HD dom... 64.7 7e-11
xla:447085 hddc2, MGC85244; HD domain containing 2; K07023 put... 62.0 4e-10
mmu:69692 Hddc2, 2310057G13Rik, MGC129436, MGC129437; HD domai... 61.2 7e-10
ath:AT2G23820 metal-dependent phosphohydrolase HD domain-conta... 61.2 8e-10
dre:554129 hddc2, fb71h08, wu:fb71h08, zgc:112330; HD domain c... 61.2 8e-10
ath:AT1G26160 metal-dependent phosphohydrolase HD domain-conta... 60.5 1e-09
sce:YGL101W Putative protein of unknown function; non-essentia... 60.1 2e-09
sce:YBR242W Putative protein of unknown function; green fluore... 57.4 1e-08
pfa:PFL1890c HD superfamily phosphohydrolase protein 34.3 0.100
ath:AT1G30600 subtilase family protein 28.9 3.8
ath:AT2G40890 CYP98A3 (cytochrome P450, family 98, subfamily A... 28.5 5.3
hsa:8881 CDC16, ANAPC6, APC6, CUT9; cell division cycle 16 hom... 28.1 6.6
eco:b2291 yfbR, ECK2285, JW2288; 5'-nucleotidase; K08722 5'-nu... 28.1 6.8
eco:b2230 yfaA, ECK2222, JW2224, pufY; conserved protein, DUF2... 27.7 9.3
> tgo:TGME49_109290 HD domain-containing protein ; K07023 putative
hydrolases of HD superfamily
Length=305
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 41/84 (48%), Positives = 51/84 (60%), Gaps = 4/84 (4%)
Query 20 LESFLSFGGKLKKLKRAGWLRYVHPSDVESVADHSF--GVAFHFLGL-PDSDLVVNGQLA 76
L +FL G+LKKLKR GW + ESVA+HSF G+ +G P S ++
Sbjct 32 LLNFLLMVGELKKLKRTGW-KLSGVRGPESVAEHSFRAGICAFLIGTDPQSSKLIRENKL 90
Query 77 DRNKCAAMALAHDLAESVVGDITP 100
DRNKC MAL HDLAE++ GDITP
Sbjct 91 DRNKCIKMALVHDLAEALAGDITP 114
> hsa:51020 HDDC2, C6orf74, MGC87330, NS5ATP2, dJ167O5.2; HD domain
containing 2; K07023 putative hydrolases of HD superfamily
Length=204
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 34/93 (36%), Positives = 52/93 (55%), Gaps = 8/93 (8%)
Query 11 TMSGEERKGLESFLSFGGKLKKLKRAGWLRYVHPSDVESVADHSFGVAFHFLGLPDSDLV 70
T SG + L FL G+LK++ R GW+ Y + ESV+DH + +A + + D L
Sbjct 8 TFSGHGARSLLQFLRLVGQLKRVPRTGWV-YRNVQRPESVSDHMYRMAVMAMVIKDDRL- 65
Query 71 VNGQLADRNKCAAMALAHDLAESVVGDITPLDG 103
++++C +AL HD+AE +VGDI P D
Sbjct 66 ------NKDRCVRLALVHDMAECIVGDIAPADN 92
> xla:447085 hddc2, MGC85244; HD domain containing 2; K07023 putative
hydrolases of HD superfamily
Length=201
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 31/86 (36%), Positives = 47/86 (54%), Gaps = 8/86 (9%)
Query 18 KGLESFLSFGGKLKKLKRAGWLRYVHPSDVESVADHSFGVAFHFLGLPDSDLVVNGQLAD 77
K L F+ G+LK++ R GW+ Y ESV+DH + +A + D L +
Sbjct 12 KSLLQFMKLVGQLKRVPRTGWI-YRQVEKPESVSDHMYRMAVMAMLTEDRKL-------N 63
Query 78 RNKCAAMALAHDLAESVVGDITPLDG 103
+++C +AL HD+AE +VGDI P D
Sbjct 64 KDRCIRLALVHDMAECIVGDIAPADN 89
> mmu:69692 Hddc2, 2310057G13Rik, MGC129436, MGC129437; HD domain
containing 2; K07023 putative hydrolases of HD superfamily
Length=199
Score = 61.2 bits (147), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 32/85 (37%), Positives = 48/85 (56%), Gaps = 8/85 (9%)
Query 19 GLESFLSFGGKLKKLKRAGWLRYVHPSDVESVADHSFGVAFHFLGLPDSDLVVNGQLADR 78
GL FL G+LK++ R GW+ Y + ESV+DH + +A + D L ++
Sbjct 11 GLLRFLRLVGQLKRVPRTGWV-YRNVEKPESVSDHMYRMAVMAMVTRDDRL-------NK 62
Query 79 NKCAAMALAHDLAESVVGDITPLDG 103
++C +AL HD+AE +VGDI P D
Sbjct 63 DRCIRLALVHDMAECIVGDIAPADN 87
> ath:AT2G23820 metal-dependent phosphohydrolase HD domain-containing
protein; K07023 putative hydrolases of HD superfamily
Length=245
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 35/79 (44%), Positives = 48/79 (60%), Gaps = 9/79 (11%)
Query 23 FLSFGGKLKKLKRAGWLRYVHPSDVESVADHSFGVAFHFLGLPDSDLV-VNGQLADRNKC 81
FLS +LK RAGW++ D ES+ADH + + + L SD+ VN R+KC
Sbjct 82 FLSLCTRLKTTPRAGWIKR-DVKDPESIADHMYRMGL--MALISSDIPGVN-----RDKC 133
Query 82 AAMALAHDLAESVVGDITP 100
MA+ HD+AE++VGDITP
Sbjct 134 MKMAIVHDIAEAIVGDITP 152
> dre:554129 hddc2, fb71h08, wu:fb71h08, zgc:112330; HD domain
containing 2; K07023 putative hydrolases of HD superfamily
Length=200
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 31/82 (37%), Positives = 47/82 (57%), Gaps = 8/82 (9%)
Query 22 SFLSFGGKLKKLKRAGWLRYVHPSDVESVADHSFGVAFHFLGLPDSDLVVNGQLADRNKC 81
F+ G+LK++ R GW+ Y + ESV+DH + ++ + L D+ VN + +C
Sbjct 6 QFMKLVGQLKRVPRTGWV-YRNIKQPESVSDHMYRMSM--MALTIQDISVN-----KERC 57
Query 82 AAMALAHDLAESVVGDITPLDG 103
+AL HDLAE +VGDI P D
Sbjct 58 MKLALVHDLAECIVGDIAPADN 79
> ath:AT1G26160 metal-dependent phosphohydrolase HD domain-containing
protein; K07023 putative hydrolases of HD superfamily
Length=258
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 34/83 (40%), Positives = 47/83 (56%), Gaps = 11/83 (13%)
Query 23 FLSFGGKLKKLKRAGWLRYVHPSDVESVADHSFGVAFHFLGLPDSDLVVNGQLA--DRNK 80
FL+ +LK KR GW+ + ES+ADH + +A L + G L DR +
Sbjct 78 FLTLCHRLKTTKRKGWINQ-GINGPESIADHMYRMALMAL--------IAGDLTGVDRER 128
Query 81 CAAMALAHDLAESVVGDITPLDG 103
C MA+ HD+AE++VGDITP DG
Sbjct 129 CIKMAIVHDIAEAIVGDITPSDG 151
> sce:YGL101W Putative protein of unknown function; non-essential
gene with similarity to YBR242W; interacts with the DNA
helicase Hpr5p; K07023 putative hydrolases of HD superfamily
Length=215
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 33/82 (40%), Positives = 50/82 (60%), Gaps = 10/82 (12%)
Query 22 SFLSFGGKLKKLKRAGWLRY-VHPSDVESVADHSFGVAFHFLGLPDSDLVVNGQLADRNK 80
+FL+ LK +R GW+ + + P ES++DH + +GL + +++ + DRNK
Sbjct 31 AFLNIIQLLKTQRRTGWVDHGIDP--CESISDHMYR-----MGL--TTMLITDKNVDRNK 81
Query 81 CAAMALAHDLAESVVGDITPLD 102
C +AL HD AES+VGDITP D
Sbjct 82 CIRIALVHDFAESLVGDITPND 103
> sce:YBR242W Putative protein of unknown function; green fluorescent
protein (GFP)-fusion protein localizes to the cytoplasm
and nucleus; YBR242W is not an essential gene; K07023 putative
hydrolases of HD superfamily
Length=238
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 30/81 (37%), Positives = 51/81 (62%), Gaps = 8/81 (9%)
Query 22 SFLSFGGKLKKLKRAGWLRYVHPSDVESVADHSFGVAFHFLGLPDSDLVVNGQLADRNKC 81
+FL+ +LK +R G+L + + ES++DH + ++ + + DS + +R+KC
Sbjct 50 AFLNVVQQLKIQRRTGYLD-LGIKECESISDHMYRLSIITMLIKDSRV-------NRDKC 101
Query 82 AAMALAHDLAESVVGDITPLD 102
+AL HD+AES+VGDITP+D
Sbjct 102 VRIALVHDIAESLVGDITPVD 122
> pfa:PFL1890c HD superfamily phosphohydrolase protein
Length=822
Score = 34.3 bits (77), Expect = 0.100, Method: Composition-based stats.
Identities = 26/73 (35%), Positives = 38/73 (52%), Gaps = 7/73 (9%)
Query 26 FGGKLKKLKRAGWLRYVHPSDVESVADHSFGVAF----HFLGLPD-SDLVVN-GQLADRN 79
F +L+ L + G +YV+P S +HS GV F +F L + S+L N G+L
Sbjct 384 FFQRLRSLSQLGACQYVYPGATHSRFEHSLGVGFLSAKYFTHLCNRSNLSPNHGELKRML 443
Query 80 KCAAMA-LAHDLA 91
+C +A L HDL
Sbjct 444 RCVQIAGLCHDLG 456
> ath:AT1G30600 subtilase family protein
Length=832
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 18/66 (27%), Positives = 30/66 (45%), Gaps = 5/66 (7%)
Query 43 HPSDVESVADHSFGVAFHFLGLPDSDL-----VVNGQLADRNKCAAMALAHDLAESVVGD 97
HPS + ++ H++ V HF G+ + + N +L A AL+ + S D
Sbjct 179 HPSFSDKISGHTYSVPPHFTGVCEVTIGFPPGSCNRKLIGARHFAESALSRGVLNSSQDD 238
Query 98 ITPLDG 103
+P DG
Sbjct 239 ASPFDG 244
> ath:AT2G40890 CYP98A3 (cytochrome P450, family 98, subfamily
A, polypeptide 3); monooxygenase/ p-coumarate 3-hydroxylase;
K09754 p-coumarate 3-hydroxylase [EC:1.14.13.-]
Length=508
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 13/49 (26%), Positives = 24/49 (48%), Gaps = 0/49 (0%)
Query 5 GSGIVKTMSGEERKGLESFLSFGGKLKKLKRAGWLRYVHPSDVESVADH 53
G+V E + + + L G L + WLR++ P+D ++ A+H
Sbjct 194 AEGVVDEQGLEFKAIVSNGLKLGASLSIAEHIPWLRWMFPADEKAFAEH 242
> hsa:8881 CDC16, ANAPC6, APC6, CUT9; cell division cycle 16 homolog
(S. cerevisiae); K03353 anaphase-promoting complex subunit
6
Length=620
Score = 28.1 bits (61), Expect = 6.6, Method: Composition-based stats.
Identities = 24/94 (25%), Positives = 38/94 (40%), Gaps = 14/94 (14%)
Query 7 GIVKTMSGEERKGLESFLSFGGKLKKLKRAGWLRYVHPSDVESVADHSFGVAFH------ 60
G M G + + +LS L+K W+ Y H VES D + F
Sbjct 306 GCYYLMVGHKNEHARRYLSKATTLEKTYGPAWIAYGHSFAVESEHDQAMAAYFTAAQLMK 365
Query 61 -------FLGLPDSDLVVNGQLADRNKCAAMALA 87
++GL + L N +LA+R A+++A
Sbjct 366 GCHLPMLYIGL-EYGLTNNSKLAERFFSQALSIA 398
> eco:b2291 yfbR, ECK2285, JW2288; 5'-nucleotidase; K08722 5'-nucleotidase
[EC:3.1.3.5]
Length=199
Score = 28.1 bits (61), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 38/80 (47%), Gaps = 4/80 (5%)
Query 23 FLSFGGKLKKLKRAGWLRYVHPSDVESVADHSFGVAFHFLGLPDSDLVVNGQLADRNKCA 82
F + +LK + R +R V E+V++HS VA L G + + A
Sbjct 6 FFAHLSRLKLINRWPLMRNVR---TENVSEHSLQVAMVAHALAAIKNRKFGGNVNAERIA 62
Query 83 AMALAHDLAESVVGDI-TPL 101
+A+ HD +E + GD+ TP+
Sbjct 63 LLAMYHDASEVLTGDLPTPV 82
> eco:b2230 yfaA, ECK2222, JW2224, pufY; conserved protein, DUF2138
family
Length=562
Score = 27.7 bits (60), Expect = 9.3, Method: Composition-based stats.
Identities = 20/75 (26%), Positives = 32/75 (42%), Gaps = 10/75 (13%)
Query 25 SFGGKLKKLKRAGWLRYVHPSDVESVADHSFGVAFHFLGLPDSDLVVNGQLADRNKCAAM 84
SF G ++ GW +V +D + D SF D V N A + C A+
Sbjct 277 SFAGVRFEMGNDGWHSFVALNDESASVDASF----------DFTPVWNSMPAGASFCVAV 326
Query 85 ALAHDLAESVVGDIT 99
+H +AE ++ I+
Sbjct 327 PYSHGIAEEMLSHIS 341
Lambda K H
0.319 0.138 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2027061836
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40