bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8028_orf1
Length=114
Score E
Sequences producing significant alignments: (Bits) Value
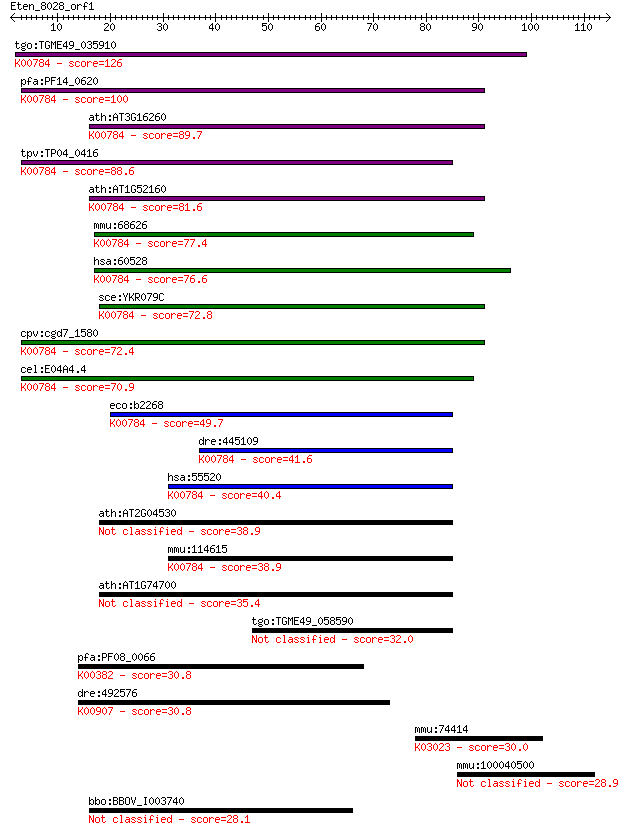
tgo:TGME49_035910 hypothetical protein ; K00784 ribonuclease Z... 126 2e-29
pfa:PF14_0620 metal-dependent hydrolase, putative (EC:3.1.26.1... 100 1e-21
ath:AT3G16260 TRZ4; TRZ4 (TRNASE Z 4); 3'-tRNA processing endo... 89.7 2e-18
tpv:TP04_0416 hypothetical protein; K00784 ribonuclease Z [EC:... 88.6 4e-18
ath:AT1G52160 TRZ3; TRZ3 (TRNASE Z 3); 3'-tRNA processing endo... 81.6 5e-16
mmu:68626 Elac2, 1110017O07Rik, D11Wsu80e, Hpc2; elaC homolog ... 77.4 1e-14
hsa:60528 ELAC2, ELC2, FLJ10530, FLJ36693, FLJ42848, HPC2; ela... 76.6 2e-14
sce:YKR079C TRZ1; Trz1p (EC:3.1.26.11); K00784 ribonuclease Z ... 72.8 2e-13
cpv:cgd7_1580 mbl domain containing protein ; K00784 ribonucle... 72.4 3e-13
cel:E04A4.4 hoe-1; Homolog Of ELAC2 (cancer susceptibility loc... 70.9 1e-12
eco:b2268 rbn, ECK2262, ecoZ, elaC, JW2263, rnz, zipD; RNase B... 49.7 2e-06
dre:445109 elac1, zgc:91956; elaC homolog 1 (E. coli) (EC:3.1.... 41.6 6e-04
hsa:55520 ELAC1, D29, FLJ59261; elaC homolog 1 (E. coli) (EC:3... 40.4 0.002
ath:AT2G04530 CPZ; CPZ; 3'-tRNA processing endoribonuclease 38.9 0.004
mmu:114615 Elac1, 2610018O07Rik, 8430417G19Rik; elaC homolog 1... 38.9 0.004
ath:AT1G74700 TRZ1; TRZ1 (TRNASE Z 1); 3'-tRNA processing endo... 35.4 0.040
tgo:TGME49_058590 hypothetical protein 32.0 0.47
pfa:PF08_0066 lipoamide dehydrogenase, putative (EC:1.8.1.4); ... 30.8 1.1
dre:492576 mylkb, im:7142009; myosin light chain kinase b; K00... 30.8 1.2
mmu:74414 Polr3c, 4933407E01Rik, RPC3, RPC62; polymerase (RNA)... 30.0 1.9
mmu:100040500 Gm2808; predicted gene 2808 28.9
bbo:BBOV_I003740 19.m02312; dopey, N-terminal domain containin... 28.1 7.8
> tgo:TGME49_035910 hypothetical protein ; K00784 ribonuclease
Z [EC:3.1.26.11]
Length=681
Score = 126 bits (316), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 62/107 (57%), Positives = 76/107 (71%), Gaps = 10/107 (9%)
Query 2 PDSFGLRVDFE----------KLSFVYSGDTRPCSQLFALAHKCTVLLHEATFEDALHSE 51
P SFGLRVDFE LS VYSGDTRPC QLF LA TVL+HEATFEDAL E
Sbjct 491 PHSFGLRVDFEIPASDANSRKDLSVVYSGDTRPCRQLFDLARNATVLIHEATFEDALIQE 550
Query 52 AVDKKHSCVSEVLKGAAECGCESVILTHFSQRYPRLPHIRFSERGPA 98
A++K+HS +SEV++GA +C C +++LTHFSQRYP++P + G A
Sbjct 551 AIEKRHSSLSEVVRGALDCRCANLVLTHFSQRYPKIPVLTLEAGGDA 597
> pfa:PF14_0620 metal-dependent hydrolase, putative (EC:3.1.26.11);
K00784 ribonuclease Z [EC:3.1.26.11]
Length=858
Score = 100 bits (249), Expect = 1e-21, Method: Composition-based stats.
Identities = 43/89 (48%), Positives = 64/89 (71%), Gaps = 1/89 (1%)
Query 3 DSFGLRVDFEKL-SFVYSGDTRPCSQLFALAHKCTVLLHEATFEDALHSEAVDKKHSCVS 61
+S+G++V+ +K+ S VYS DTRPC + + C +L+HEATF+D L EA++KKHS
Sbjct 723 ESYGIKVENKKIGSIVYSADTRPCENVKKFSKNCDILIHEATFDDELLGEAINKKHSTTK 782
Query 62 EVLKGAAECGCESVILTHFSQRYPRLPHI 90
E + + E C+++ILTHFSQRYP++P I
Sbjct 783 EAMDISLEVQCKTLILTHFSQRYPKVPKI 811
> ath:AT3G16260 TRZ4; TRZ4 (TRNASE Z 4); 3'-tRNA processing endoribonuclease/
catalytic; K00784 ribonuclease Z [EC:3.1.26.11]
Length=942
Score = 89.7 bits (221), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 41/75 (54%), Positives = 50/75 (66%), Gaps = 0/75 (0%)
Query 16 FVYSGDTRPCSQLFALAHKCTVLLHEATFEDALHSEAVDKKHSCVSEVLKGAAECGCESV 75
VYSGDTRPC ++ + TVL+HEATFEDAL EAV K HS E +K + G
Sbjct 802 MVYSGDTRPCPEMVEASKGATVLIHEATFEDALVEEAVAKNHSTTKEAIKVGSSAGVYRT 861
Query 76 ILTHFSQRYPRLPHI 90
+LTHFSQRYP++P I
Sbjct 862 VLTHFSQRYPKIPVI 876
> tpv:TP04_0416 hypothetical protein; K00784 ribonuclease Z [EC:3.1.26.11]
Length=500
Score = 88.6 bits (218), Expect = 4e-18, Method: Composition-based stats.
Identities = 39/82 (47%), Positives = 53/82 (64%), Gaps = 0/82 (0%)
Query 3 DSFGLRVDFEKLSFVYSGDTRPCSQLFALAHKCTVLLHEATFEDALHSEAVDKKHSCVSE 62
DS+G+R+D+E S VYSGDT+PC L L+HEATF D A+ HS VS+
Sbjct 399 DSYGIRIDYEFWSLVYSGDTKPCDNLIKHCENVNFLIHEATFTDEYAENAMKTLHSTVSQ 458
Query 63 VLKGAAECGCESVILTHFSQRY 84
++ AA+C E++ LTHFSQR+
Sbjct 459 AIEVAAQCNVETLFLTHFSQRF 480
> ath:AT1G52160 TRZ3; TRZ3 (TRNASE Z 3); 3'-tRNA processing endoribonuclease/
catalytic; K00784 ribonuclease Z [EC:3.1.26.11]
Length=890
Score = 81.6 bits (200), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 36/75 (48%), Positives = 48/75 (64%), Gaps = 0/75 (0%)
Query 16 FVYSGDTRPCSQLFALAHKCTVLLHEATFEDALHSEAVDKKHSCVSEVLKGAAECGCESV 75
VYSGD+RPC + + T+L+HEATFEDAL EA+ K HS E + + +
Sbjct 757 MVYSGDSRPCPETVEASRDATILIHEATFEDALIEEALAKNHSTTKEAIDVGSAANVYRI 816
Query 76 ILTHFSQRYPRLPHI 90
+LTHFSQRYP++P I
Sbjct 817 VLTHFSQRYPKIPVI 831
> mmu:68626 Elac2, 1110017O07Rik, D11Wsu80e, Hpc2; elaC homolog
2 (E. coli) (EC:3.1.26.11); K00784 ribonuclease Z [EC:3.1.26.11]
Length=831
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 34/72 (47%), Positives = 44/72 (61%), Gaps = 0/72 (0%)
Query 17 VYSGDTRPCSQLFALAHKCTVLLHEATFEDALHSEAVDKKHSCVSEVLKGAAECGCESVI 76
VYSGDT PC L + T+L+HEAT ED L EAV+K HS S+ + E ++
Sbjct 658 VYSGDTMPCEALVQMGKDATLLIHEATLEDGLEEEAVEKTHSTTSQAINVGMRMNAEFIM 717
Query 77 LTHFSQRYPRLP 88
L HFSQRY ++P
Sbjct 718 LNHFSQRYAKIP 729
> hsa:60528 ELAC2, ELC2, FLJ10530, FLJ36693, FLJ42848, HPC2; elaC
homolog 2 (E. coli) (EC:3.1.26.11); K00784 ribonuclease
Z [EC:3.1.26.11]
Length=786
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 37/81 (45%), Positives = 48/81 (59%), Gaps = 2/81 (2%)
Query 17 VYSGDTRPCSQLFALAHKCTVLLHEATFEDALHSEAVDKKHSCVSEVLKGAAECGCESVI 76
VYSGDT PC L + T+L+HEAT ED L EAV+K HS S+ + E ++
Sbjct 622 VYSGDTMPCEALVRMGKDATLLIHEATLEDGLEEEAVEKTHSTTSQAISVGMRMNAEFIM 681
Query 77 LTHFSQRYPRLPHI--RFSER 95
L HFSQRY ++P FSE+
Sbjct 682 LNHFSQRYAKVPLFSPNFSEK 702
> sce:YKR079C TRZ1; Trz1p (EC:3.1.26.11); K00784 ribonuclease
Z [EC:3.1.26.11]
Length=838
Score = 72.8 bits (177), Expect = 2e-13, Method: Composition-based stats.
Identities = 34/75 (45%), Positives = 49/75 (65%), Gaps = 2/75 (2%)
Query 18 YSGDTRPCSQLFAL--AHKCTVLLHEATFEDALHSEAVDKKHSCVSEVLKGAAECGCESV 75
YSGDTRP + F+L + +L+HEAT E+ L +AV KKH ++E + + + +
Sbjct 696 YSGDTRPNIEKFSLEIGYNSDLLIHEATLENQLLEDAVKKKHCTINEAIGVSNKMNARKL 755
Query 76 ILTHFSQRYPRLPHI 90
ILTHFSQRYP+LP +
Sbjct 756 ILTHFSQRYPKLPQL 770
> cpv:cgd7_1580 mbl domain containing protein ; K00784 ribonuclease
Z [EC:3.1.26.11]
Length=821
Score = 72.4 bits (176), Expect = 3e-13, Method: Composition-based stats.
Identities = 35/89 (39%), Positives = 53/89 (59%), Gaps = 1/89 (1%)
Query 3 DSFGLRVDFEKLSFVYSGDT-RPCSQLFALAHKCTVLLHEATFEDALHSEAVDKKHSCVS 61
+S +R + V+SGDT PC+ L + C +L+HEATFED+L +A +K HS +S
Sbjct 683 NSLKIRDGYSLFKIVFSGDTATPCTSLEYASKDCDILIHEATFEDSLSKDAQEKNHSTIS 742
Query 62 EVLKGAAECGCESVILTHFSQRYPRLPHI 90
+ A + ++LTHFSQRY +P +
Sbjct 743 GAIGTAYNSLAKFLLLTHFSQRYFSMPKV 771
> cel:E04A4.4 hoe-1; Homolog Of ELAC2 (cancer susceptibility locus)
family member (hoe-1); K00784 ribonuclease Z [EC:3.1.26.11]
Length=833
Score = 70.9 bits (172), Expect = 1e-12, Method: Composition-based stats.
Identities = 33/86 (38%), Positives = 53/86 (61%), Gaps = 2/86 (2%)
Query 3 DSFGLRVDFEKLSFVYSGDTRPCSQLFALAHKCTVLLHEATFEDALHSEAVDKKHSCVSE 62
+ F +RV +++ V+SGDT+PC L VL+HE+TFED ++A+ K+HS + +
Sbjct 681 NGFVMRVAGKRI--VFSGDTKPCDLLVEEGKDADVLVHESTFEDGHEADAMRKRHSTMGQ 738
Query 63 VLKGAAECGCESVILTHFSQRYPRLP 88
+ + +ILTHFS RYP++P
Sbjct 739 AVDVGKRMNAKHIILTHFSARYPKVP 764
> eco:b2268 rbn, ECK2262, ecoZ, elaC, JW2263, rnz, zipD; RNase
BN, tRNA processing enzyme; K00784 ribonuclease Z [EC:3.1.26.11]
Length=305
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 34/65 (52%), Gaps = 0/65 (0%)
Query 20 GDTRPCSQLFALAHKCTVLLHEATFEDALHSEAVDKKHSCVSEVLKGAAECGCESVILTH 79
GDT PC LA V++HEAT + + ++A + HS + A E G +I+TH
Sbjct 211 GDTGPCDAALDLAKGVDVMVHEATLDITMEAKANSRGHSSTRQAATLAREAGVGKLIITH 270
Query 80 FSQRY 84
S RY
Sbjct 271 VSSRY 275
> dre:445109 elac1, zgc:91956; elaC homolog 1 (E. coli) (EC:3.1.26.11);
K00784 ribonuclease Z [EC:3.1.26.11]
Length=372
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 28/48 (58%), Gaps = 0/48 (0%)
Query 37 VLLHEATFEDALHSEAVDKKHSCVSEVLKGAAECGCESVILTHFSQRY 84
VL+HEAT E+ +AV+ HS A C ++++L HFSQRY
Sbjct 274 VLVHEATLENGQQEKAVEHGHSTPGMAAAVALTCEAKTLVLHHFSQRY 321
> hsa:55520 ELAC1, D29, FLJ59261; elaC homolog 1 (E. coli) (EC:3.1.26.11);
K00784 ribonuclease Z [EC:3.1.26.11]
Length=363
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 30/54 (55%), Gaps = 0/54 (0%)
Query 31 LAHKCTVLLHEATFEDALHSEAVDKKHSCVSEVLKGAAECGCESVILTHFSQRY 84
L + +L+HEAT +DA +A + HS A C + ++LTHFSQRY
Sbjct 265 LCFEADLLIHEATLDDAQMDKAKEHGHSTPQMAATFAKLCRAKRLVLTHFSQRY 318
> ath:AT2G04530 CPZ; CPZ; 3'-tRNA processing endoribonuclease
Length=354
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 42/74 (56%), Gaps = 9/74 (12%)
Query 18 YSGDTRPCSQL---FALAHKCTVLLHEATFED----ALHSEAVDKKHSCVSEVLKGAAEC 70
++GDT L A A + VL+ EATF D H++A+ H+ +S++++ A
Sbjct 251 FTGDTTSEYMLDPRNADALRAKVLITEATFLDESFSTEHAQALG--HTHISQIIENAKWI 308
Query 71 GCESVILTHFSQRY 84
++V+LTHFS RY
Sbjct 309 RSKTVLLTHFSSRY 322
> mmu:114615 Elac1, 2610018O07Rik, 8430417G19Rik; elaC homolog
1 (E. coli) (EC:3.1.26.11); K00784 ribonuclease Z [EC:3.1.26.11]
Length=362
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 30/54 (55%), Gaps = 0/54 (0%)
Query 31 LAHKCTVLLHEATFEDALHSEAVDKKHSCVSEVLKGAAECGCESVILTHFSQRY 84
L + +L+HEAT +D+ +A + HS A C + ++LTHFSQRY
Sbjct 264 LCFEADLLIHEATLDDSQMDKAREHGHSTPQMAAAFAKLCRAKRLVLTHFSQRY 317
> ath:AT1G74700 TRZ1; TRZ1 (TRNASE Z 1); 3'-tRNA processing endoribonuclease
Length=280
Score = 35.4 bits (80), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 41/72 (56%), Gaps = 5/72 (6%)
Query 18 YSGDTRP---CSQLFALAHKCTVLLHEATF-EDALHSE-AVDKKHSCVSEVLKGAAECGC 72
++GDT + A A K VL+ E+TF +D++ E A D H +SE++ A +
Sbjct 182 FTGDTTSDFVVDETNADALKAKVLVMESTFLDDSVSVEHARDYGHIHISEIVNHAEKFEN 241
Query 73 ESVILTHFSQRY 84
++++L HFS RY
Sbjct 242 KAILLIHFSARY 253
> tgo:TGME49_058590 hypothetical protein
Length=460
Score = 32.0 bits (71), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 47 ALHSEAVDKKHSCVSEVLKGAAECGCESVILTHFSQRY 84
AL EA + HS + A G + ++LTHFSQR+
Sbjct 403 ALDEEAFARGHSTAAMAGDFAGRAGAQRLVLTHFSQRF 440
> pfa:PF08_0066 lipoamide dehydrogenase, putative (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=666
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 27/59 (45%), Gaps = 5/59 (8%)
Query 14 LSFVYSGDTRPCSQLFALAHKCTVLLHEATFEDALHSEAVD-----KKHSCVSEVLKGA 67
+ +Y DT+ +F + + +VL+HEA L A D H VSEVL A
Sbjct 598 VKIIYKEDTKEILGMFIVGNYASVLIHEAVLAINLKLSAFDLAYMVHSHPTVSEVLDTA 656
> dre:492576 mylkb, im:7142009; myosin light chain kinase b; K00907
myosin-light-chain kinase [EC:2.7.11.18]
Length=1002
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 17/62 (27%), Positives = 32/62 (51%), Gaps = 10/62 (16%)
Query 14 LSFVYSGDTRPCSQLFALAH---KCTVLLHEATFEDALHSEAVDKKHSCVSEVLKGAAEC 70
++++ G T S+ L+H KC++ + +A ED + +++C +E G AEC
Sbjct 197 ITWILDGKTVKPSKFIVLSHEGEKCSLTIDKALPED-------EGRYTCRAENAHGKAEC 249
Query 71 GC 72
C
Sbjct 250 SC 251
> mmu:74414 Polr3c, 4933407E01Rik, RPC3, RPC62; polymerase (RNA)
III (DNA directed) polypeptide C; K03023 DNA-directed RNA
polymerase III subunit RPC3
Length=533
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 15/24 (62%), Gaps = 0/24 (0%)
Query 78 THFSQRYPRLPHIRFSERGPANPS 101
THF QR P +P S+RGP P+
Sbjct 154 THFVQRCPLVPDTDSSDRGPPPPA 177
> mmu:100040500 Gm2808; predicted gene 2808
Length=1336
Score = 28.9 bits (63), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 15/26 (57%), Gaps = 0/26 (0%)
Query 86 RLPHIRFSERGPANPSENPKGAAKGA 111
++PH+ S R P P PKG A GA
Sbjct 790 KMPHLADSSRAPLQPLAKPKGGAAGA 815
> bbo:BBOV_I003740 19.m02312; dopey, N-terminal domain containing
protein
Length=2138
Score = 28.1 bits (61), Expect = 7.8, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 20/50 (40%), Gaps = 0/50 (0%)
Query 16 FVYSGDTRPCSQLFALAHKCTVLLHEATFEDALHSEAVDKKHSCVSEVLK 65
F Y GD PCS L+ L + T + A KH C S +K
Sbjct 1330 FNYEGDLPPCSLSSILSLAANGLANHETNDSARQLFGFGNKHKCTSATIK 1379
Lambda K H
0.318 0.134 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2047611720
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40