bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8037_orf1
Length=186
Score E
Sequences producing significant alignments: (Bits) Value
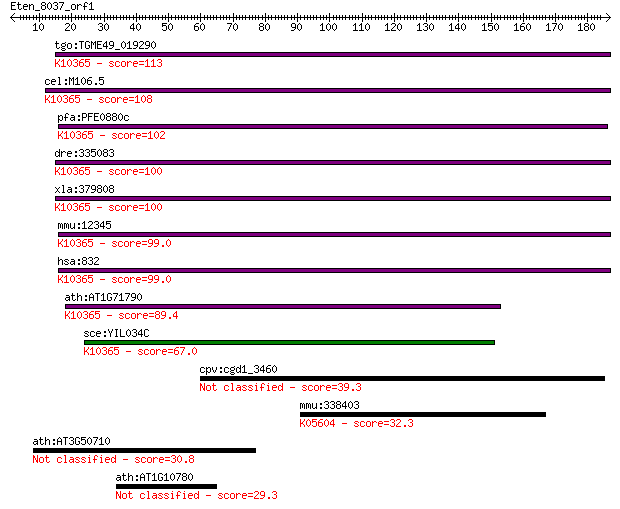
tgo:TGME49_019290 F-actin capping protein beta subunit, putati... 113 3e-25
cel:M106.5 cap-2; CAP-z protein family member (cap-2); K10365 ... 108 9e-24
pfa:PFE0880c F-actin capping protein beta subunit, putative; K... 102 1e-21
dre:335083 capzb, wu:fk65f06, zgc:66149; capping protein (acti... 100 2e-21
xla:379808 capzb, MGC52754, capb, cappb; capping protein (acti... 100 4e-21
mmu:12345 Capzb, 1700120C01Rik, AI325129, CPB1, CPB2, CPbeat2,... 99.0 8e-21
hsa:832 CAPZB, CAPB, CAPPB, CAPZ, MGC104401, MGC129749, MGC129... 99.0 9e-21
ath:AT1G71790 F-actin capping protein beta subunit family prot... 89.4 6e-18
sce:YIL034C CAP2; Beta subunit of the capping protein (CP) het... 67.0 4e-11
cpv:cgd1_3460 F-actin capping protein, beta subunit 39.3 0.009
mmu:338403 Cndp1, AI746433, Cn1; carnosine dipeptidase 1 (meta... 32.3 1.0
ath:AT3G50710 F-box family protein 30.8 2.6
ath:AT1G10780 F-box family protein 29.3 9.0
> tgo:TGME49_019290 F-actin capping protein beta subunit, putative
; K10365 capping protein (actin filament) muscle Z-line,
beta
Length=329
Score = 113 bits (283), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 70/185 (37%), Positives = 102/185 (55%), Gaps = 14/185 (7%)
Query 15 KETLELAAALVRLGPLDAVNESLAHAAALCPSIEGLLRDRCQGIPELVVDPAAGKYFIAS 74
+E+ A +L R P ++ ++A LCP + L R L D A K+FIA
Sbjct 14 EESWIAAVSLTRRMPPKLIDRTVAGVQHLCPDLSIQLLTRVDRRLRLCFDSEARKFFIAC 73
Query 75 YFNKLKNSFRSPWTDTWLPPISDAS----------KPAPHLRQLEQQFNAAYNSYRDAHY 124
+N+ +SFRSPWT+ ++ +D S KPA +LR LE +N ++SYR A+Y
Sbjct 74 IYNQHGSSFRSPWTNAYIEG-NDVSVLGAVPQRRIKPADNLRNLETTYNRIFDSYRRAYY 132
Query 125 GGGVSSVYAWPLQSQDGFAAAFLLLK--EAPVDEPLLPLCFFT-HRLEVTLTATNAHYKL 181
GGVSSVY W L S+DGFA AFL+ + V C+ + H +EVT + +N HY+L
Sbjct 133 EGGVSSVYLWSLPSEDGFAGAFLVRHALDGGVSGDGPRGCWESVHVVEVTQSTSNVHYRL 192
Query 182 HSTLL 186
ST++
Sbjct 193 TSTVM 197
> cel:M106.5 cap-2; CAP-z protein family member (cap-2); K10365
capping protein (actin filament) muscle Z-line, beta
Length=270
Score = 108 bits (271), Expect = 9e-24, Method: Compositional matrix adjust.
Identities = 61/175 (34%), Positives = 96/175 (54%), Gaps = 3/175 (1%)
Query 12 GQQKETLELAAALVRLGPLDAVNESLAHAAALCPSIEGLLRDRCQGIPELVVDPAAGKYF 71
G+Q+ L+ A L+R P +++L LCP + L ++ D GK +
Sbjct 2 GEQQ--LDCALDLMRRLPPQHCDKNLTDLIDLCPHLVDDLLSTIDQPLKIAADRETGKQY 59
Query 72 IASYFNKLKNSFRSPWTDTWLPPISDASKPAPHLRQLEQQFNAAYNSYRDAHYGGGVSSV 131
+ +N+ +S+RSPW++T+ PP+ D P+ R++E + NAA+ SYRD ++ GGVSSV
Sbjct 60 LLCDYNRDGDSYRSPWSNTYDPPLEDGQLPSEKRRKMEIEANAAFESYRDLYFEGGVSSV 119
Query 132 YAWPLQSQDGFAAAFLLLKEAPVDEPLLPLCFFTHRLEVTLTATNAHYKLHSTLL 186
Y W L + GFA L+ KE + + H +E+T A AHYKL ST++
Sbjct 120 YFWDLDNG-GFAGIVLIKKEGDGAKNITGCWDSIHVIEITERARQAHYKLTSTIM 173
> pfa:PFE0880c F-actin capping protein beta subunit, putative;
K10365 capping protein (actin filament) muscle Z-line, beta
Length=292
Score = 102 bits (253), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 53/171 (30%), Positives = 91/171 (53%), Gaps = 1/171 (0%)
Query 16 ETLELAAALVRLGPLDAVNESLAHAAALCPSIEGLLRDRCQGIPELVVDPAAGKYFIASY 75
+ +E A + P ++++ + + SI + +G ++ D KY++ +
Sbjct 4 DKMEAALNICNTLPGHVFDDTIKMLSRIDQSITNNILINKEGSIKINYDKEENKYYLGNM 63
Query 76 FNKLKNSFRSPWTDTWLPP-ISDASKPAPHLRQLEQQFNAAYNSYRDAHYGGGVSSVYAW 134
FNK K+S+RSP+T+ + P ++ P HLR LE +N ++ YR A+Y G+SSVY W
Sbjct 64 FNKEKDSYRSPYTNIYFPEHYINSYVPPEHLRTLEILYNKIFDRYRKAYYMNGLSSVYLW 123
Query 135 PLQSQDGFAAAFLLLKEAPVDEPLLPLCFFTHRLEVTLTATNAHYKLHSTL 185
P +DGF A FL+ K+ D+ TH ++V +T N HY++ T+
Sbjct 124 PNPIEDGFVACFLIKKKEIFDKETNIKWEATHLIQVNITNLNVHYQISCTI 174
> dre:335083 capzb, wu:fk65f06, zgc:66149; capping protein (actin
filament) muscle Z-line, beta; K10365 capping protein (actin
filament) muscle Z-line, beta
Length=273
Score = 100 bits (250), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 60/175 (34%), Positives = 97/175 (55%), Gaps = 6/175 (3%)
Query 15 KETLELAAALVRLGPLDAVNESLAHAAALCPSI-EGLLRDRCQGIPELVVDPAAGKYFIA 73
++ L+ A L+R P + ++L+ L PS+ E LL Q + ++ D GK ++
Sbjct 3 EQQLDCALDLMRRLPPQQIEKNLSDLIDLVPSLCEDLLSSVDQPL-KIARDKVVGKDYLL 61
Query 74 SYFNKLKNSFRSPWTDTWLPPISDASKPAPHLRQLEQQFNAAYNSYRDAHYGGGVSSVYA 133
+N+ +S+RSPW++ + PPI D + P+ LR+LE + N A++ YRD ++ GGVSSVY
Sbjct 62 CDYNRDGDSYRSPWSNKYEPPIDDGAMPSARLRKLEVEANNAFDQYRDLYFEGGVSSVYL 121
Query 134 WPLQSQDGFAAAFLLLKEAPVDEPLLPLCFFTHRLEVTLTATN--AHYKLHSTLL 186
W L GFA L+ K + + H +EV ++ AHYKL ST++
Sbjct 122 WDLDH--GFAGVILIKKAGDGSKKIKGCWDSIHVVEVQEKSSGRTAHYKLTSTVM 174
> xla:379808 capzb, MGC52754, capb, cappb; capping protein (actin
filament) muscle Z-line, beta; K10365 capping protein (actin
filament) muscle Z-line, beta
Length=272
Score = 100 bits (248), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 60/175 (34%), Positives = 97/175 (55%), Gaps = 6/175 (3%)
Query 15 KETLELAAALVRLGPLDAVNESLAHAAALCPSI-EGLLRDRCQGIPELVVDPAAGKYFIA 73
++ L+ A L+R P + ++L+ L PS+ E LL Q + ++ D GK ++
Sbjct 3 EQQLDCALDLMRRLPPQQIEKNLSDLIDLVPSLCEDLLSSVDQPL-KIARDKVLGKDYLL 61
Query 74 SYFNKLKNSFRSPWTDTWLPPISDASKPAPHLRQLEQQFNAAYNSYRDAHYGGGVSSVYA 133
+N+ +S+RSPW++ + PP+ D + P+ LR+LE + N A++ YRD +Y GGVSSVY
Sbjct 62 CDYNRDGDSYRSPWSNKYDPPLEDGAMPSARLRKLEVEANNAFDQYRDLYYEGGVSSVYL 121
Query 134 WPLQSQDGFAAAFLLLKEAPVDEPLLPLCFFTHRLEVTLTATN--AHYKLHSTLL 186
W L GFA L+ K + + H +EV ++ AHYKL ST++
Sbjct 122 WDLDH--GFAGVILIKKAGDGSKKIKGCWDSIHVVEVQEKSSGRTAHYKLTSTVM 174
> mmu:12345 Capzb, 1700120C01Rik, AI325129, CPB1, CPB2, CPbeat2,
CPbeta1, CPbeta2, Cappb1; capping protein (actin filament)
muscle Z-line, beta; K10365 capping protein (actin filament)
muscle Z-line, beta
Length=277
Score = 99.0 bits (245), Expect = 8e-21, Method: Compositional matrix adjust.
Identities = 59/174 (33%), Positives = 96/174 (55%), Gaps = 6/174 (3%)
Query 16 ETLELAAALVRLGPLDAVNESLAHAAALCPSI-EGLLRDRCQGIPELVVDPAAGKYFIAS 74
+ L+ A L+R P + ++L+ L PS+ E LL Q + ++ D GK ++
Sbjct 4 QQLDCALDLMRRLPPQQIEKNLSDLIDLVPSLCEDLLSSVDQPL-KIARDKVVGKDYLLC 62
Query 75 YFNKLKNSFRSPWTDTWLPPISDASKPAPHLRQLEQQFNAAYNSYRDAHYGGGVSSVYAW 134
+N+ +S+RSPW++ + PP+ D + P+ LR+LE + N A++ YRD ++ GGVSSVY W
Sbjct 63 DYNRDGDSYRSPWSNKYDPPLEDGAMPSARLRKLEVEANNAFDQYRDLYFEGGVSSVYLW 122
Query 135 PLQSQDGFAAAFLLLKEAPVDEPLLPLCFFTHRLEVTLTAT--NAHYKLHSTLL 186
L GFA L+ K + + H +EV ++ AHYKL ST++
Sbjct 123 DLDH--GFAGVILIKKAGDGSKKIKGCWDSIHVVEVQEKSSGRTAHYKLTSTVM 174
> hsa:832 CAPZB, CAPB, CAPPB, CAPZ, MGC104401, MGC129749, MGC129750;
capping protein (actin filament) muscle Z-line, beta;
K10365 capping protein (actin filament) muscle Z-line, beta
Length=272
Score = 99.0 bits (245), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 59/174 (33%), Positives = 96/174 (55%), Gaps = 6/174 (3%)
Query 16 ETLELAAALVRLGPLDAVNESLAHAAALCPSI-EGLLRDRCQGIPELVVDPAAGKYFIAS 74
+ L+ A L+R P + ++L+ L PS+ E LL Q + ++ D GK ++
Sbjct 4 QQLDCALDLMRRLPPQQIEKNLSDLIDLVPSLCEDLLSSVDQPL-KIARDKVVGKDYLLC 62
Query 75 YFNKLKNSFRSPWTDTWLPPISDASKPAPHLRQLEQQFNAAYNSYRDAHYGGGVSSVYAW 134
+N+ +S+RSPW++ + PP+ D + P+ LR+LE + N A++ YRD ++ GGVSSVY W
Sbjct 63 DYNRDGDSYRSPWSNKYDPPLEDGAMPSARLRKLEVEANNAFDQYRDLYFEGGVSSVYLW 122
Query 135 PLQSQDGFAAAFLLLKEAPVDEPLLPLCFFTHRLEVTLTATN--AHYKLHSTLL 186
L GFA L+ K + + H +EV ++ AHYKL ST++
Sbjct 123 DLDH--GFAGVILIKKAGDGSKKIKGCWDSIHVVEVQEKSSGRTAHYKLTSTVM 174
> ath:AT1G71790 F-actin capping protein beta subunit family protein;
K10365 capping protein (actin filament) muscle Z-line,
beta
Length=256
Score = 89.4 bits (220), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 48/135 (35%), Positives = 75/135 (55%), Gaps = 1/135 (0%)
Query 18 LELAAALVRLGPLDAVNESLAHAAALCPSIEGLLRDRCQGIPELVVDPAAGKYFIASYFN 77
+E A L+R P +L+ +L P L + +++ D +GK FI +N
Sbjct 1 MEAALGLLRRMPPKQSETALSALLSLIPQHSSDLLSQVDLPLQVLRDIESGKDFILCEYN 60
Query 78 KLKNSFRSPWTDTWLPPISDASKPAPHLRQLEQQFNAAYNSYRDAHYGGGVSSVYAWPLQ 137
+ +S+RSPW++ +LPP+ DA P+ LR+LE + N + YRD +Y GG+SSVY W
Sbjct 61 RDADSYRSPWSNKYLPPLEDALYPSSELRKLEVEANDIFAIYRDQYYEGGISSVYMWE-D 119
Query 138 SQDGFAAAFLLLKEA 152
+GF A FL+ K+
Sbjct 120 DNEGFVACFLIKKDG 134
> sce:YIL034C CAP2; Beta subunit of the capping protein (CP) heterodimer
(Cap1p and Cap2p) which binds to the barbed ends
of actin filaments preventing further polymerization; localized
predominantly to cortical actin patches; K10365 capping
protein (actin filament) muscle Z-line, beta
Length=287
Score = 67.0 bits (162), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 45/137 (32%), Positives = 70/137 (51%), Gaps = 11/137 (8%)
Query 24 LVRLGPLDAVNESLAHAAALCPSIEGLLRDRCQGIPELVVDPA-AGKYFIASYFNKLKNS 82
L RL P + E+L + L P++ L D A + + ++ +N+ +S
Sbjct 13 LRRLNPT-TLQENLNNLIELQPNLAQDLLSSVDVPLSTQKDSADSNREYLCCDYNRDIDS 71
Query 83 FRSPWTDTWLPPIS-----DASKPAPHLRQLEQQFNAAYNSYRDAHYGGGVSSVYAWPLQ 137
FRSPW++T+ P +S D+ P+ LR+LE N +++ YRD +Y GG+SSVY W L
Sbjct 72 FRSPWSNTYYPELSPKDLQDSPFPSAPLRKLEILANDSFDVYRDLYYEGGISSVYLWDLN 131
Query 138 SQD----GFAAAFLLLK 150
+D FA L K
Sbjct 132 EEDFNGHDFAGVVLFKK 148
> cpv:cgd1_3460 F-actin capping protein, beta subunit
Length=319
Score = 39.3 bits (90), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 38/171 (22%), Positives = 69/171 (40%), Gaps = 47/171 (27%)
Query 60 ELVVDPAAGKYFIASYFNKLKNSFRSPWT-DTWLPPISD--------------ASKPAPH 104
++++D Y++ N+++N FRSP+T ++ P S A+ H
Sbjct 22 KVILDLETKMYYLGCNSNRIENYFRSPYTYKFYIDPKSKEEYDSEKAESFKEIANNDLNH 81
Query 105 LRQLEQQFNAAYNSY--RDAHYGGG-----------VSSVYAWPLQSQDGFAAAFLL--- 148
L+ LE +F Y Y + GG +S+VY + L+ D F F++
Sbjct 82 LKMLETEFQKVYEIYCQNYTYINGGLEEDICDHGVLLSNVYCYDLEG-DSFGTCFVMKHI 140
Query 149 ----------LKEAPVDEPLLPLCFF---THRLEVTLTATN--AHYKLHST 184
+ E L + +F H +E L+ ++ A Y++ ST
Sbjct 141 INPFNFLNDNINEKSSTNDLTEIVYFLDIIHNVETVLSHSSGIATYRVGST 191
> mmu:338403 Cndp1, AI746433, Cn1; carnosine dipeptidase 1 (metallopeptidase
M20 family) (EC:3.4.13.20); K05604 beta-Ala-His
dipeptidase [EC:3.4.13.20]
Length=492
Score = 32.3 bits (72), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 26/79 (32%), Positives = 36/79 (45%), Gaps = 5/79 (6%)
Query 91 WLPPISDASKPAPHLRQ-LEQQFNAAYNSYRDAHYGGGVSSVYAWPLQSQDGFAAAF--L 147
W+ SD+ +P P LRQ L Q A + R+ G GV S+ Q DG + +
Sbjct 31 WVAIESDSVQPVPRLRQKLFQMMALAADKLRN--LGAGVESIDLGSQQMPDGQSLPIPPI 88
Query 148 LLKEAPVDEPLLPLCFFTH 166
LL E D +CF+ H
Sbjct 89 LLAELGSDPEKPTVCFYGH 107
> ath:AT3G50710 F-box family protein
Length=427
Score = 30.8 bits (68), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 19/74 (25%), Positives = 36/74 (48%), Gaps = 5/74 (6%)
Query 8 ALYSGQQKETLELAAALVRLGPLDAVNESLAHAAALCPSIEGLLRDR-----CQGIPELV 62
+LY+ ++ E L+LA+ +V P+D SL +C + R C + ELV
Sbjct 127 SLYTYEKLEVLKLASTVVLNVPIDVCFPSLKSLHLVCVEYKTKKSHRRLLSGCPVLEELV 186
Query 63 VDPAAGKYFIASYF 76
+D + + + S++
Sbjct 187 LDKSYNSFHVRSFY 200
> ath:AT1G10780 F-box family protein
Length=418
Score = 29.3 bits (64), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 11/31 (35%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 34 NESLAHAAALCPSIEGLLRDRCQGIPELVVD 64
+ SL+HA CP++ LL C+G+ + +D
Sbjct 163 DSSLSHALRACPNLSNLLLLACEGVKSISID 193
Lambda K H
0.319 0.134 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5170784960
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40