bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8074_orf1
Length=79
Score E
Sequences producing significant alignments: (Bits) Value
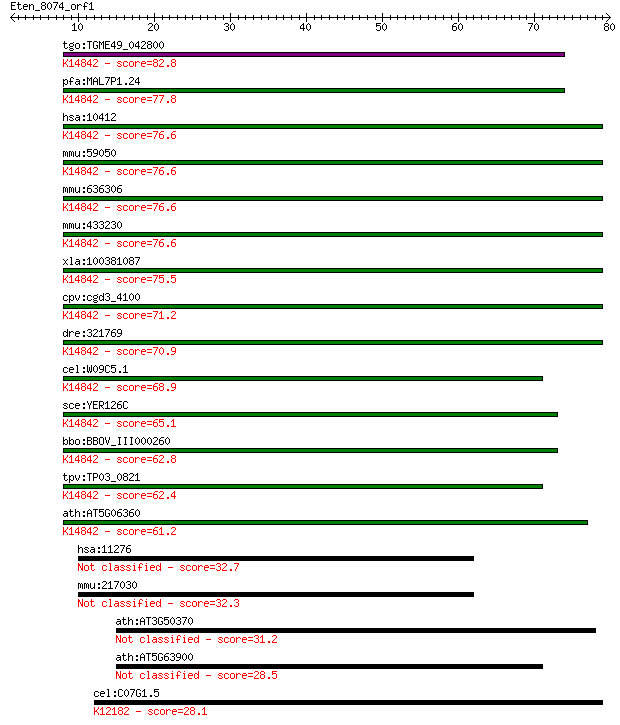
tgo:TGME49_042800 TGF-beta-inducible nuclear protein 1, putati... 82.8 2e-16
pfa:MAL7P1.24 cdk105, putative; K14842 ribosome biogenesis pro... 77.8 9e-15
hsa:10412 NSA2, CDK105, HCL-G1, HCLG1, HUSSY29, TINP1, YR-29; ... 76.6 2e-14
mmu:59050 Nsa2, 5730427N09Rik, LNR42, MGC118054, Nsa2p, Tinp1,... 76.6 2e-14
mmu:636306 ribosome biogenesis protein NSA2 homolog; K14842 ri... 76.6 2e-14
mmu:433230 Gm5515, EG433230; predicted gene 5515; K14842 ribos... 76.6 2e-14
xla:100381087 nsa2, cdk105, hcl-g1, hclg1, hussy-29, hussy29, ... 75.5 4e-14
cpv:cgd3_4100 conserved protein, COG SSU ribosomal protein S8E... 71.2 8e-13
dre:321769 nsa2, fb34b10, wu:fb34b10, zgc:56334; NSA2 ribosome... 70.9 1e-12
cel:W09C5.1 hypothetical protein; K14842 ribosome biogenesis p... 68.9 3e-12
sce:YER126C NSA2; Nsa2p; K14842 ribosome biogenesis protein NSA2 65.1 5e-11
bbo:BBOV_III000260 17.m07048; ribosomal protein Se8; K14842 ri... 62.8 3e-10
tpv:TP03_0821 hypothetical protein; K14842 ribosome biogenesis... 62.4 3e-10
ath:AT5G06360 ribosomal protein S8e family protein; K14842 rib... 61.2 8e-10
hsa:11276 SYNRG, AP1GBP1, FLJ34482, MGC104959, SYNG; synergin,... 32.7 0.26
mmu:217030 Synrg, AF007009, Ap1gbp1, C76297, L71-5, SYNG; syne... 32.3 0.36
ath:AT3G50370 hypothetical protein 31.2 0.86
ath:AT5G63900 PHD finger family protein 28.5 5.6
cel:C07G1.5 hgrs-1; Hepatocyte Growth factor-Regulated TK Subs... 28.1 7.6
> tgo:TGME49_042800 TGF-beta-inducible nuclear protein 1, putative
; K14842 ribosome biogenesis protein NSA2
Length=260
Score = 82.8 bits (203), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 41/66 (62%), Positives = 54/66 (81%), Gaps = 0/66 (0%)
Query 8 MPQNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQK 67
MPQNEYIE H+KRFG R D ER+RKK AREPH+ +++A+ LRGIK+K+ NKKR++EK
Sbjct 1 MPQNEYIELHQKRFGRRLDHHERQRKKAAREPHKLSKQARRLRGIKSKLFNKKRYVEKAT 60
Query 68 MKKRIQ 73
MKK ++
Sbjct 61 MKKTLK 66
> pfa:MAL7P1.24 cdk105, putative; K14842 ribosome biogenesis protein
NSA2
Length=259
Score = 77.8 bits (190), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 38/66 (57%), Positives = 52/66 (78%), Gaps = 0/66 (0%)
Query 8 MPQNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQK 67
MPQN+YIE H+KR+G RFD E+ RKKEAR+ H+ + +AK LRGIKAK+ NKK++ EK
Sbjct 1 MPQNDYIELHRKRYGYRFDHFEKERKKEARKVHKDSLKAKKLRGIKAKIYNKKKYTEKVN 60
Query 68 MKKRIQ 73
+KK ++
Sbjct 61 LKKTLK 66
> hsa:10412 NSA2, CDK105, HCL-G1, HCLG1, HUSSY29, TINP1, YR-29;
NSA2 ribosome biogenesis homolog (S. cerevisiae); K14842 ribosome
biogenesis protein NSA2
Length=260
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 38/71 (53%), Positives = 57/71 (80%), Gaps = 0/71 (0%)
Query 8 MPQNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQK 67
MPQNEYIE H+KR+G R D E++RKKE+RE HER+++AK + G+KAK+ +K+R EK +
Sbjct 1 MPQNEYIELHRKRYGYRLDYHEKKRKKESREAHERSKKAKKMIGLKAKLYHKQRHAEKIQ 60
Query 68 MKKRIQLEDRK 78
MKK I++ +++
Sbjct 61 MKKTIKMHEKR 71
> mmu:59050 Nsa2, 5730427N09Rik, LNR42, MGC118054, Nsa2p, Tinp1,
Yr29; NSA2 ribosome biogenesis homolog (S. cerevisiae); K14842
ribosome biogenesis protein NSA2
Length=260
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 38/71 (53%), Positives = 56/71 (78%), Gaps = 0/71 (0%)
Query 8 MPQNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQK 67
MPQNEYIE H+KR+G R D E++RKKE RE HER+++AK + G+KAK+ +K+R EK +
Sbjct 1 MPQNEYIELHRKRYGYRLDYHEKKRKKEGREAHERSKKAKKMIGLKAKLYHKQRHAEKIQ 60
Query 68 MKKRIQLEDRK 78
MKK I++ +++
Sbjct 61 MKKTIKMHEKR 71
> mmu:636306 ribosome biogenesis protein NSA2 homolog; K14842
ribosome biogenesis protein NSA2
Length=260
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 38/71 (53%), Positives = 56/71 (78%), Gaps = 0/71 (0%)
Query 8 MPQNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQK 67
MPQNEYIE H+KR+G R D E++RKKE RE HER+++AK + G+KAK+ +K+R EK +
Sbjct 1 MPQNEYIELHRKRYGYRLDYHEKKRKKEGREAHERSKKAKKMIGLKAKLYHKQRHAEKIQ 60
Query 68 MKKRIQLEDRK 78
MKK I++ +++
Sbjct 61 MKKTIKMHEKR 71
> mmu:433230 Gm5515, EG433230; predicted gene 5515; K14842 ribosome
biogenesis protein NSA2
Length=260
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 38/71 (53%), Positives = 56/71 (78%), Gaps = 0/71 (0%)
Query 8 MPQNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQK 67
MPQNEYIE H+KR+G R D E++RKKE RE HER+++AK + G+KAK+ +K+R EK +
Sbjct 1 MPQNEYIELHRKRYGYRLDYHEKKRKKEGREAHERSKKAKKMIGLKAKLYHKQRHAEKIQ 60
Query 68 MKKRIQLEDRK 78
MKK I++ +++
Sbjct 61 MKKTIKMHEKR 71
> xla:100381087 nsa2, cdk105, hcl-g1, hclg1, hussy-29, hussy29,
tinp1, yr-29; NSA2 ribosome biogenesis homolog; K14842 ribosome
biogenesis protein NSA2
Length=260
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 37/71 (52%), Positives = 57/71 (80%), Gaps = 0/71 (0%)
Query 8 MPQNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQK 67
MPQN+YIE H+KR+G R D E++RKKE+RE HER+++AK L G+KAK+ +K+R EK +
Sbjct 1 MPQNDYIELHRKRYGYRLDYHEKKRKKESREAHERSQKAKKLIGLKAKLYHKQRHAEKIQ 60
Query 68 MKKRIQLEDRK 78
MKK +++ +++
Sbjct 61 MKKTLKMHEKR 71
> cpv:cgd3_4100 conserved protein, COG SSU ribosomal protein S8E
; K14842 ribosome biogenesis protein NSA2
Length=260
Score = 71.2 bits (173), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 37/71 (52%), Positives = 53/71 (74%), Gaps = 0/71 (0%)
Query 8 MPQNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQK 67
MPQNEYIE H+KR G RFD E++RK E+R+ H+ ++ A+ LRGIKAK+ NKKR+ EK
Sbjct 1 MPQNEYIELHRKRHGYRFDHFEKKRKFESRKVHKDSQHAQKLRGIKAKLYNKKRYAEKAT 60
Query 68 MKKRIQLEDRK 78
++K ++ + K
Sbjct 61 LRKLVKANEEK 71
> dre:321769 nsa2, fb34b10, wu:fb34b10, zgc:56334; NSA2 ribosome
biogenesis homolog (S. cerevisiae); K14842 ribosome biogenesis
protein NSA2
Length=260
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 36/71 (50%), Positives = 55/71 (77%), Gaps = 0/71 (0%)
Query 8 MPQNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQK 67
MPQNE+IE H+KR G R D E++RKKE+RE HER+ +A+ + G+KAK+ +K+R EK +
Sbjct 1 MPQNEHIELHRKRHGYRLDHHEKKRKKESREAHERSHKARKMIGLKAKLYHKQRHAEKIQ 60
Query 68 MKKRIQLEDRK 78
MKK I++ +++
Sbjct 61 MKKTIKMHEQR 71
> cel:W09C5.1 hypothetical protein; K14842 ribosome biogenesis
protein NSA2
Length=259
Score = 68.9 bits (167), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 37/63 (58%), Positives = 48/63 (76%), Gaps = 0/63 (0%)
Query 8 MPQNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQK 67
MPQNE+IE H+KR G R D ER+RKK AR H+R++ AK LRG KAK+ +KKR+ EK +
Sbjct 1 MPQNEHIELHRKRHGRRLDHEERQRKKLARAAHDRSQMAKTLRGHKAKLYHKKRYSEKVE 60
Query 68 MKK 70
M+K
Sbjct 61 MRK 63
> sce:YER126C NSA2; Nsa2p; K14842 ribosome biogenesis protein
NSA2
Length=261
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 35/65 (53%), Positives = 47/65 (72%), Gaps = 0/65 (0%)
Query 8 MPQNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQK 67
MPQN+YIE+H K+ G+R D ER+RK+EARE H+ + RA+ L G K K KKR+ EK
Sbjct 1 MPQNDYIERHIKQHGKRLDHEERKRKREARESHKISERAQKLTGWKGKQFAKKRYAEKVS 60
Query 68 MKKRI 72
M+K+I
Sbjct 61 MRKKI 65
> bbo:BBOV_III000260 17.m07048; ribosomal protein Se8; K14842
ribosome biogenesis protein NSA2
Length=259
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 33/65 (50%), Positives = 45/65 (69%), Gaps = 0/65 (0%)
Query 8 MPQNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQK 67
MPQNEYIE+H G RFD R RKK AR H +++ + +RG+KAK+ K+R+ EK +
Sbjct 1 MPQNEYIERHIALHGRRFDHETRMRKKTARAAHTLSKKMQKMRGLKAKIFRKRRYAEKIE 60
Query 68 MKKRI 72
MKK+I
Sbjct 61 MKKQI 65
> tpv:TP03_0821 hypothetical protein; K14842 ribosome biogenesis
protein NSA2
Length=259
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 33/63 (52%), Positives = 43/63 (68%), Gaps = 0/63 (0%)
Query 8 MPQNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQK 67
MPQNEYIE+H K G RFD + RKK AR P ++RAK LRGIK+K+ K ++ EK +
Sbjct 1 MPQNEYIERHIKLHGRRFDHLTKTRKKAARAPLRESKRAKTLRGIKSKIFKKHKYAEKIQ 60
Query 68 MKK 70
+K
Sbjct 61 FRK 63
> ath:AT5G06360 ribosomal protein S8e family protein; K14842 ribosome
biogenesis protein NSA2
Length=260
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 33/69 (47%), Positives = 46/69 (66%), Gaps = 0/69 (0%)
Query 8 MPQNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQK 67
MPQ +YI+ H+KR G R D ER+RKKEARE H+ + A+ GIK K+ KK + EK
Sbjct 1 MPQGDYIDLHRKRNGYRLDHFERKRKKEAREVHKHSTMAQKSLGIKGKMIAKKNYAEKAL 60
Query 68 MKKRIQLED 76
MKK +++ +
Sbjct 61 MKKTLKMHE 69
> hsa:11276 SYNRG, AP1GBP1, FLJ34482, MGC104959, SYNG; synergin,
gamma
Length=1236
Score = 32.7 bits (73), Expect = 0.26, Method: Composition-based stats.
Identities = 16/52 (30%), Positives = 29/52 (55%), Gaps = 2/52 (3%)
Query 10 QNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKR 61
Q ++ E+ +KRF ++ E RK+ R+ E+ ++ +LL +K K K R
Sbjct 116 QKQFAEEQQKRFEQQQKLLEEERKR--RQFEEQKQKLRLLSSVKPKTGEKSR 165
> mmu:217030 Synrg, AF007009, Ap1gbp1, C76297, L71-5, SYNG; synergin,
gamma
Length=1138
Score = 32.3 bits (72), Expect = 0.36, Method: Composition-based stats.
Identities = 16/52 (30%), Positives = 29/52 (55%), Gaps = 2/52 (3%)
Query 10 QNEYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKR 61
Q ++ E+ +KRF ++ E RK+ R+ E+ ++ +LL +K K K R
Sbjct 113 QKQFAEEQQKRFEQQQKLLEEERKR--RQFEEQKQKLRLLSSVKPKTGEKNR 162
> ath:AT3G50370 hypothetical protein
Length=2156
Score = 31.2 bits (69), Expect = 0.86, Method: Composition-based stats.
Identities = 26/77 (33%), Positives = 41/77 (53%), Gaps = 16/77 (20%)
Query 15 EQHKKRFGERFDAAERRRKKEAREPH--------ERARRAKLLRGIKAKVENKKR-FLEK 65
E+ + R D +RR ++EARE E RRA+ LR K+K E K R F+E+
Sbjct 475 EEERLRLAREQDERQRRLEEEAREAAFRNEQERLEATRRAEELR--KSKEEEKHRLFMEE 532
Query 66 QKMK-----KRIQLEDR 77
++ K K ++LE++
Sbjct 533 ERRKQAAKQKLLELEEK 549
> ath:AT5G63900 PHD finger family protein
Length=557
Score = 28.5 bits (62), Expect = 5.6, Method: Composition-based stats.
Identities = 15/56 (26%), Positives = 29/56 (51%), Gaps = 0/56 (0%)
Query 15 EQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQKMKK 70
+Q K R G D ++KK + E RR K+++ +K ++ ++ +K K +K
Sbjct 185 DQEKIRIGLNVDVIHEQQKKRRKMASEEIRRPKMVKSLKKVLQVMEKKQQKNKHEK 240
> cel:C07G1.5 hgrs-1; Hepatocyte Growth factor-Regulated TK Substrate
(HRS) family member (hgrs-1); K12182 hepatocyte growth
factor-regulated tyrosine kinase substrate
Length=729
Score = 28.1 bits (61), Expect = 7.6, Method: Composition-based stats.
Identities = 18/67 (26%), Positives = 32/67 (47%), Gaps = 5/67 (7%)
Query 12 EYIEQHKKRFGERFDAAERRRKKEAREPHERARRAKLLRGIKAKVENKKRFLEKQKMKKR 71
E ++ H GE R+ E RE HER R+ ++ + + ++ LE +MKK
Sbjct 464 ESLQDHLANIGE-----ARQAIDEMREEHERKRQERMAEEQRLRQAQMQQTLEMMRMKKH 518
Query 72 IQLEDRK 78
L +++
Sbjct 519 AMLMEQR 525
Lambda K H
0.323 0.138 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2063098576
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40