bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
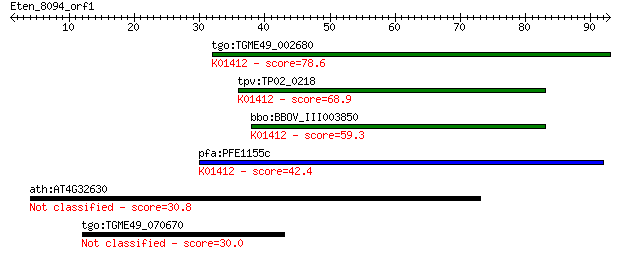
Query= Eten_8094_orf1
Length=92
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_002680 mitochondrial-processing peptidase alpha sub... 78.6 5e-15
tpv:TP02_0218 ubiquinol-cytochrome C reductase complex core pr... 68.9 4e-12
bbo:BBOV_III003850 17.m07356; mitochondrial processing peptida... 59.3 3e-09
pfa:PFE1155c mitochondrial processing peptidase alpha subunit,... 42.4 4e-04
ath:AT4G32630 ARF GTPase activator/ zinc ion binding 30.8 1.1
tgo:TGME49_070670 hypothetical protein 30.0 1.7
> tgo:TGME49_002680 mitochondrial-processing peptidase alpha subunit,
putative (EC:3.4.24.64); K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=563
Score = 78.6 bits (192), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 33/61 (54%), Positives = 44/61 (72%), Gaps = 0/61 (0%)
Query 32 AGQQPQYRRVPFVREDLSLVMQEVPDFKYYYIGNEAPSAYPYKDVPMDQPILNPDEFPEY 91
AG P+YRRVPFV+ED+ VM+EVP+FKYYY+G E Y+ +P+DQ IL P + +Y
Sbjct 65 AGPAPEYRRVPFVKEDMEKVMEEVPEFKYYYVGKENTKGNVYEGIPLDQSILEPADLRDY 124
Query 92 V 92
V
Sbjct 125 V 125
> tpv:TP02_0218 ubiquinol-cytochrome C reductase complex core
protein II, mitochondrial precursor (EC:1.10.2.2); K01412 mitochondrial
processing peptidase [EC:3.4.24.64]
Length=525
Score = 68.9 bits (167), Expect = 4e-12, Method: Composition-based stats.
Identities = 26/47 (55%), Positives = 35/47 (74%), Gaps = 0/47 (0%)
Query 36 PQYRRVPFVREDLSLVMQEVPDFKYYYIGNEAPSAYPYKDVPMDQPI 82
P Y++VPF++EDL V+ EVP+F +YY G+ YPYKDVPM +PI
Sbjct 31 PDYKQVPFLKEDLDSVLSEVPEFNFYYFGSNTDEDYPYKDVPMTEPI 77
> bbo:BBOV_III003850 17.m07356; mitochondrial processing peptidase
alpha subunit; K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=496
Score = 59.3 bits (142), Expect = 3e-09, Method: Composition-based stats.
Identities = 24/45 (53%), Positives = 32/45 (71%), Gaps = 0/45 (0%)
Query 38 YRRVPFVREDLSLVMQEVPDFKYYYIGNEAPSAYPYKDVPMDQPI 82
Y +VPFV EDL VM+EVPDF++YYIG + PY +PM++ I
Sbjct 8 YEKVPFVDEDLERVMEEVPDFRFYYIGRDDGGRNPYSSIPMNEEI 52
> pfa:PFE1155c mitochondrial processing peptidase alpha subunit,
putative; K01412 mitochondrial processing peptidase [EC:3.4.24.64]
Length=534
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 21/62 (33%), Positives = 36/62 (58%), Gaps = 3/62 (4%)
Query 30 VAAGQQPQYRRVPFVREDLSLVMQEVPDFKYYYIGNEAPSAYPYKDVPMDQPILNPDEFP 89
+ G++ + +V F +E + V++EV F YYY NE YKD+P++ I+N +FP
Sbjct 36 IYTGEKQNFNKVSFKKEKIEDVIKEV-KFDYYYF-NEGKKN-KYKDIPLNISIINESDFP 92
Query 90 EY 91
+
Sbjct 93 PF 94
> ath:AT4G32630 ARF GTPase activator/ zinc ion binding
Length=628
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 22/70 (31%), Positives = 32/70 (45%), Gaps = 1/70 (1%)
Query 4 LSASQRPL-ATAAPPQTTATAAPTAAAVAAGQQPQYRRVPFVREDLSLVMQEVPDFKYYY 62
L SQ P ATA P + A A+ + GQQ Q P R++ S + E F +
Sbjct 544 LMTSQSPFHATALSPNSPALASHLSPGALMGQQSQVNMSPSFRQEYSGLGTEGNTFNGVH 603
Query 63 IGNEAPSAYP 72
++A + YP
Sbjct 604 TFHQANNGYP 613
> tgo:TGME49_070670 hypothetical protein
Length=1320
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 12 ATAAPPQTTATAAPTAAAVAAGQQPQYRRVP 42
T AP TT+ A+P AA + +G +P R++P
Sbjct 114 GTDAPGDTTSVASPPAARLGSGGKPWGRKIP 144
Lambda K H
0.314 0.130 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2003222032
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40