bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8111_orf1
Length=113
Score E
Sequences producing significant alignments: (Bits) Value
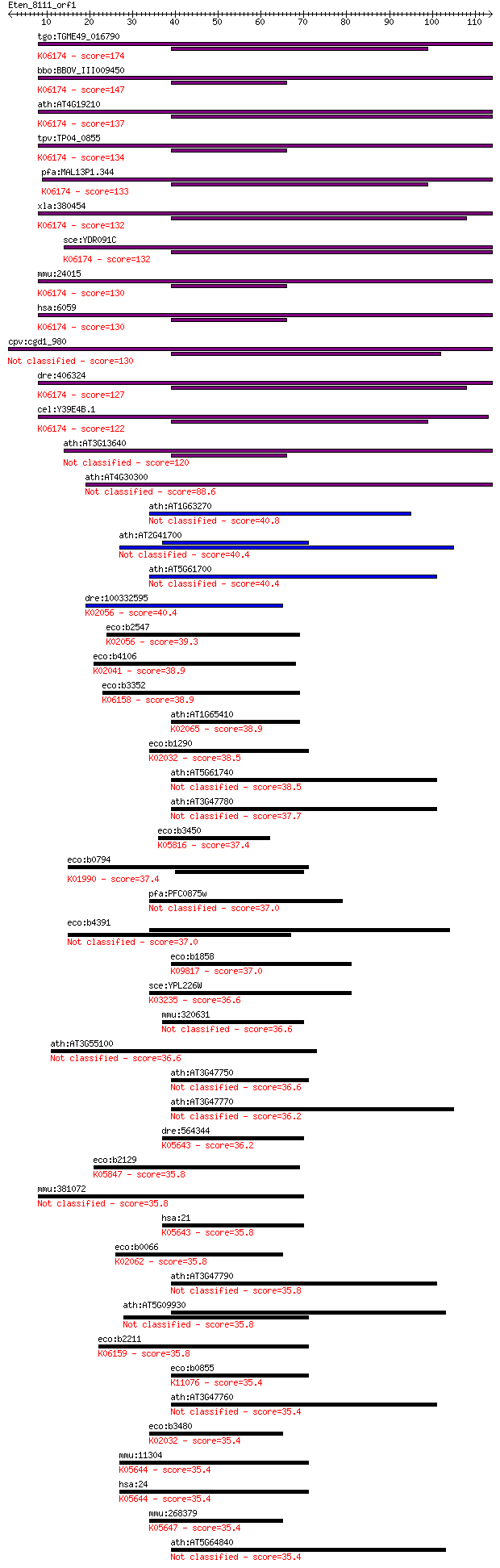
tgo:TGME49_016790 ABC transporter, putative (EC:1.2.7.8 3.6.3.... 174 6e-44
bbo:BBOV_III009450 17.m07819; ABC transporter, ATP-binding dom... 147 9e-36
ath:AT4G19210 ATRLI2; ATRLI2; transporter; K06174 ATP-binding ... 137 8e-33
tpv:TP04_0855 RNAse L inhibitor; K06174 ATP-binding cassette, ... 134 6e-32
pfa:MAL13P1.344 RNAse L inhibitor protein, putative; K06174 AT... 133 2e-31
xla:380454 abce1, MGC53437; ATP-binding cassette, sub-family E... 132 3e-31
sce:YDR091C RLI1; Essential iron-sulfur protein required for r... 132 3e-31
mmu:24015 Abce1, C79080, Oabp, RLI, RNS41, Rnaseli; ATP-bindin... 130 9e-31
hsa:6059 ABCE1, ABC38, OABP, RLI, RNASEL1, RNASELI, RNS4I; ATP... 130 9e-31
cpv:cgd1_980 RNase L inhibitor-like protein 130 1e-30
dre:406324 abce1, MGC56045, wu:fb34c09, wu:fe47b01, wu:fi09g07... 127 1e-29
cel:Y39E4B.1 abce-1; ABC transporter, class E family member (a... 122 4e-28
ath:AT3G13640 ATRLI1; ATRLI1; transporter 120 1e-27
ath:AT4G30300 ATNAP15; transporter 88.6 4e-18
ath:AT1G63270 ATNAP10; transporter 40.8 0.001
ath:AT2G41700 ATPase, coupled to transmembrane movement of sub... 40.4 0.001
ath:AT5G61700 ATATH16; ATPase, coupled to transmembrane moveme... 40.4 0.002
dre:100332595 sugar abc transporter, putative-like; K02056 sim... 40.4 0.002
eco:b2547 yphE, ECK2544, JW2531; fused predicted sugar transpo... 39.3 0.003
eco:b4106 phnC, ECK4099, JW4067; phosphonate/organophosphate e... 38.9 0.004
eco:b3352 yheS, ECK3340, JW3315; fused predicted transporter s... 38.9 0.004
ath:AT1G65410 ATNAP11 (ARABIDOPSIS THALIANA NON-INTRINSIC ABC ... 38.9 0.004
eco:b1290 sapF, ECK1285, JW1283; antimicrobial peptide transpo... 38.5 0.005
ath:AT5G61740 ATATH14; ATPase, coupled to transmembrane moveme... 38.5 0.006
ath:AT3G47780 ATATH6; ATATH6; ATPase, coupled to transmembrane... 37.7 0.008
eco:b3450 ugpC, ECK3434, JW3415; glycerol-3-phosphate transpor... 37.4 0.011
eco:b0794 ybhF, ECK0783, JW5104; fused predicted transporter s... 37.4 0.011
pfa:PFC0875w ABC transporter, putative 37.0 0.014
eco:b4391 yjjK, ECK4383, JW4354; fused predicted transporter s... 37.0 0.016
eco:b1858 znuC, ECK1859, JW1847, yebM; zinc transporter subuni... 37.0 0.017
sce:YPL226W NEW1; ATP binding cassette protein that cosediment... 36.6 0.018
mmu:320631 Abca15, 4930500I12Rik; ATP-binding cassette, sub-fa... 36.6 0.020
ath:AT3G55100 ABC transporter family protein 36.6 0.020
ath:AT3G47750 ABCA4; ABCA4 (ATP BINDING CASSETTE SUBFAMILY A4)... 36.6 0.021
ath:AT3G47770 ATATH5; ATATH5; ATPase, coupled to transmembrane... 36.2 0.026
dre:564344 ABC Transporter family member (abt-4)-like; K05643 ... 36.2 0.028
eco:b2129 yehX, ECK2122, JW2117; predicted transporter subunit... 35.8 0.030
mmu:381072 Abca17; ATP-binding cassette, sub-family A (ABC1), ... 35.8 0.031
hsa:21 ABCA3, ABC-C, ABC3, EST111653, LBM180, MGC166979, MGC72... 35.8 0.031
eco:b0066 thiQ, ECK0067, JW0065, sfuC, yabJ; thiamin transport... 35.8 0.032
ath:AT3G47790 ATATH7; ATATH7; ATPase, coupled to transmembrane... 35.8 0.032
ath:AT5G09930 ATGCN2; ATGCN2; transporter 35.8 0.035
eco:b2211 yojI, ECK2203, JW2199, yojJ; Microcin J25 efflux pum... 35.8 0.036
eco:b0855 potG, ECK0846, JW5818; putrescine transporter subuni... 35.4 0.041
ath:AT3G47760 ATATH4; ATATH4; ATPase, coupled to transmembrane... 35.4 0.041
eco:b3480 nikE, ECK3464, hydC, hydD, JW3445; nickel transporte... 35.4 0.042
mmu:11304 Abca4, AW050280, Abc10, Abcr, D430003I15Rik, RmP; AT... 35.4 0.042
hsa:24 ABCA4, ABC10, ABCR, ARMD2, CORD3, DKFZp781N1972, FFM, F... 35.4 0.045
mmu:268379 Abca13, 9830132L24, A930002G16Rik, AI956815; ATP-bi... 35.4 0.049
ath:AT5G64840 ATGCN5; ATGCN5 (A. THALIANA GENERAL CONTROL NON-... 35.4 0.049
> tgo:TGME49_016790 ABC transporter, putative (EC:1.2.7.8 3.6.3.24
3.6.3.25); K06174 ATP-binding cassette, sub-family E, member
1
Length=613
Score = 174 bits (441), Expect = 6e-44, Method: Compositional matrix adjust.
Identities = 79/106 (74%), Positives = 93/106 (87%), Gaps = 0/106 (0%)
Query 8 VQRLHFYEYPALTKTLGTFKLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNS 67
+QRLHFY+YPA+TKTLG+FKL VE G FSDSEI+VMLGQNGTGKST IRMLAGLL D
Sbjct 355 IQRLHFYKYPAMTKTLGSFKLRVESGHFSDSEILVMLGQNGTGKSTLIRMLAGLLKADEE 414
Query 68 EALPSFAVSYKPQIVTAKFQGTVKELFYAKIRDAFNHPQFQTDVVR 113
LP+ VSYKPQ +TAK+QGTV++LF+AKIR++FNHPQFQTDVV+
Sbjct 415 VELPNLHVSYKPQTITAKYQGTVRDLFFAKIRESFNHPQFQTDVVK 460
Score = 28.1 bits (61), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 35/60 (58%), Gaps = 4/60 (6%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPDNSEALPSFAVSYKPQIVTAKFQGTVKELFYAKI 98
+++ ++G NG GKST +++L+ L P+ L ++ Q + A F+G+ + F+ ++
Sbjct 115 QVLGLVGTNGIGKSTALKILSAKLKPN----LGKYSNPPDWQEILAFFRGSELQNFFTRM 170
> bbo:BBOV_III009450 17.m07819; ABC transporter, ATP-binding domain
containing protein; K06174 ATP-binding cassette, sub-family
E, member 1
Length=616
Score = 147 bits (371), Expect = 9e-36, Method: Compositional matrix adjust.
Identities = 65/109 (59%), Positives = 87/109 (79%), Gaps = 3/109 (2%)
Query 8 VQRLHFYEYPALTKTLGTFKLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNS 67
++RLH Y+YP + KTLGTF L V G F DSEI+VMLG+NGTGK+TFI+MLAG+L+ DN+
Sbjct 356 IERLHNYKYPEMKKTLGTFSLTVSAGDFCDSEILVMLGENGTGKTTFIKMLAGILAADNA 415
Query 68 ---EALPSFAVSYKPQIVTAKFQGTVKELFYAKIRDAFNHPQFQTDVVR 113
EA+P ++SYKPQI+TAKF GT+++L KI++AF P FQTDV++
Sbjct 416 EAGEAMPKLSISYKPQIITAKFDGTLRQLLMMKIKEAFGSPMFQTDVIK 464
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 12/27 (44%), Positives = 21/27 (77%), Gaps = 0/27 (0%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPD 65
+I+ ++G NG GKST +++L+G L P+
Sbjct 118 QILGLVGTNGIGKSTALKILSGKLKPN 144
> ath:AT4G19210 ATRLI2; ATRLI2; transporter; K06174 ATP-binding
cassette, sub-family E, member 1
Length=605
Score = 137 bits (345), Expect = 8e-33, Method: Compositional matrix adjust.
Identities = 61/110 (55%), Positives = 84/110 (76%), Gaps = 4/110 (3%)
Query 8 VQRLHFYEYPALTKTLGTFKLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNS 67
+Q Y+YP +TKT G F+L V +G F+DS+I+VMLG+NGTGK+TFIRMLAGLL PD++
Sbjct 344 IQSYARYKYPTMTKTQGNFRLRVSEGEFTDSQIIVMLGENGTGKTTFIRMLAGLLKPDDT 403
Query 68 EA----LPSFAVSYKPQIVTAKFQGTVKELFYAKIRDAFNHPQFQTDVVR 113
E +P F VSYKPQ ++ KFQ +V+ L + KIRD++ HPQF +DV++
Sbjct 404 EGPDREIPEFNVSYKPQKISPKFQNSVRHLLHQKIRDSYMHPQFMSDVMK 453
Score = 33.1 bits (74), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 41/79 (51%), Gaps = 8/79 (10%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPDNSEALPSFAVSYKPQIVTAKFQGTVKELFYAKI 98
+++ ++G NG GKST +++LAG L P+ L F Q + F+G+ + ++ +I
Sbjct 104 QVLGLVGTNGIGKSTALKILAGKLKPN----LGRFTSPPDWQEILTHFRGSELQNYFTRI 159
Query 99 RD----AFNHPQFQTDVVR 113
+ A PQ+ + R
Sbjct 160 LEDNLKAIIKPQYVDHIPR 178
> tpv:TP04_0855 RNAse L inhibitor; K06174 ATP-binding cassette,
sub-family E, member 1
Length=636
Score = 134 bits (338), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 61/109 (55%), Positives = 84/109 (77%), Gaps = 3/109 (2%)
Query 8 VQRLHFYEYPALTKTLGTFKLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNS 67
++ LH Y+YP L K LG+F L V G F+DSEI+V+LG+NGTGK+TFI+MLAG L PDN+
Sbjct 355 IESLHCYKYPQLDKKLGSFSLTVMPGDFNDSEIIVLLGENGTGKTTFIKMLAGKLQPDNA 414
Query 68 EA---LPSFAVSYKPQIVTAKFQGTVKELFYAKIRDAFNHPQFQTDVVR 113
E +P +VSYKPQ ++ KF GT+++LF++KIR++F P FQ DVV+
Sbjct 415 ECEDLMPKLSVSYKPQKLSVKFDGTLRQLFHSKIRESFLCPIFQADVVK 463
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 11/27 (40%), Positives = 21/27 (77%), Gaps = 0/27 (0%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPD 65
+++ ++G NG GKST +++L+G L P+
Sbjct 117 QVLGLVGTNGIGKSTALKVLSGKLKPN 143
> pfa:MAL13P1.344 RNAse L inhibitor protein, putative; K06174
ATP-binding cassette, sub-family E, member 1
Length=619
Score = 133 bits (334), Expect = 2e-31, Method: Composition-based stats.
Identities = 58/108 (53%), Positives = 81/108 (75%), Gaps = 3/108 (2%)
Query 9 QRLHFYEYPALTKTLGTFKLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNSE 68
+RLHFY YP + KTL +F L ++KG FS+SEI V+LGQNG+GKSTFIR+ AGL+ PDN E
Sbjct 360 KRLHFYNYPTMVKTLNSFSLTIDKGHFSESEIFVLLGQNGSGKSTFIRLFAGLIKPDNLE 419
Query 69 ALP---SFAVSYKPQIVTAKFQGTVKELFYAKIRDAFNHPQFQTDVVR 113
+L S +VSYKPQ + AKF GTV++L +K++ +N P F ++++
Sbjct 420 SLSFLESLSVSYKPQQIQAKFTGTVRQLLMSKLKGLYNDPYFNNEIIK 467
Score = 30.8 bits (68), Expect = 1.0, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 35/60 (58%), Gaps = 4/60 (6%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPDNSEALPSFAVSYKPQIVTAKFQGTVKELFYAKI 98
+I+ ++G NG GKST +++L+ L P+ L F + + + + F+G ++F+ K+
Sbjct 118 QILGLVGTNGIGKSTALKILSSKLKPN----LGKFNNPPEWRDILSFFRGNELQIFFTKL 173
> xla:380454 abce1, MGC53437; ATP-binding cassette, sub-family
E (OABP), member 1; K06174 ATP-binding cassette, sub-family
E, member 1
Length=599
Score = 132 bits (332), Expect = 3e-31, Method: Composition-based stats.
Identities = 57/106 (53%), Positives = 79/106 (74%), Gaps = 0/106 (0%)
Query 8 VQRLHFYEYPALTKTLGTFKLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNS 67
++++ Y+YP + K LG F+L + G F+DSEI+VMLG+NGTGK+TFIRMLAG L PD
Sbjct 342 IKKMCMYKYPCMRKLLGDFQLRIVAGEFTDSEIMVMLGENGTGKTTFIRMLAGRLKPDEG 401
Query 68 EALPSFAVSYKPQIVTAKFQGTVKELFYAKIRDAFNHPQFQTDVVR 113
+P VSYKPQ ++ K G+V++L + KIRDA+ HPQF TDV++
Sbjct 402 GDVPVLNVSYKPQKISPKASGSVRQLLHEKIRDAYTHPQFVTDVMK 447
Score = 31.2 bits (69), Expect = 0.74, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 38/73 (52%), Gaps = 8/73 (10%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPDNSEALPSFAVSYKPQIVTAKFQGTVKELFYAKI 98
E++ ++G NG GKST +++LAG P+ L + Q + F+G+ + ++ KI
Sbjct 104 EVLGLVGTNGIGKSTALKILAGKQKPN----LGKYDDPPDWQEILTYFRGSELQNYFTKI 159
Query 99 RD----AFNHPQF 107
+ A PQ+
Sbjct 160 LEDDLKAIIKPQY 172
> sce:YDR091C RLI1; Essential iron-sulfur protein required for
ribosome biogenesis and translation initiation; facilitates
binding of a multifactor complex (MFC) of translation initiation
factors to the small ribosomal subunit; predicted ABC
family ATPase; K06174 ATP-binding cassette, sub-family E, member
1
Length=608
Score = 132 bits (332), Expect = 3e-31, Method: Composition-based stats.
Identities = 59/100 (59%), Positives = 76/100 (76%), Gaps = 0/100 (0%)
Query 14 YEYPALTKTLGTFKLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNSEALPSF 73
+ YP+L KT G F LNVE+G FSDSEI+VM+G+NGTGK+T I++LAG L PD + +P
Sbjct 354 FSYPSLKKTQGDFVLNVEEGEFSDSEILVMMGENGTGKTTLIKLLAGALKPDEGQDIPKL 413
Query 74 AVSYKPQIVTAKFQGTVKELFYAKIRDAFNHPQFQTDVVR 113
VS KPQ + KF GTV++LF+ KIR F +PQFQTDVV+
Sbjct 414 NVSMKPQKIAPKFPGTVRQLFFKKIRGQFLNPQFQTDVVK 453
Score = 32.0 bits (71), Expect = 0.56, Method: Composition-based stats.
Identities = 21/79 (26%), Positives = 42/79 (53%), Gaps = 8/79 (10%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPDNSEALPSFAVSYKPQIVTAKFQGTVKELFYAKI 98
+++ ++G NG GKST +++LAG P+ L F + Q + F+G+ + ++ K+
Sbjct 104 QVLGLVGTNGIGKSTALKILAGKQKPN----LGRFDDPPEWQEIIKYFRGSELQNYFTKM 159
Query 99 RD----AFNHPQFQTDVVR 113
+ A PQ+ ++ R
Sbjct 160 LEDDIKAIIKPQYVDNIPR 178
> mmu:24015 Abce1, C79080, Oabp, RLI, RNS41, Rnaseli; ATP-binding
cassette, sub-family E (OABP), member 1; K06174 ATP-binding
cassette, sub-family E, member 1
Length=599
Score = 130 bits (327), Expect = 9e-31, Method: Compositional matrix adjust.
Identities = 57/106 (53%), Positives = 79/106 (74%), Gaps = 0/106 (0%)
Query 8 VQRLHFYEYPALTKTLGTFKLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNS 67
V+++ Y+YP + K +G F+L + G F+DSEI+VMLG+NGTGK+TFIRMLAG L PD
Sbjct 342 VKKMCMYKYPGMKKKMGEFELAIVAGEFTDSEIMVMLGENGTGKTTFIRMLAGRLKPDEG 401
Query 68 EALPSFAVSYKPQIVTAKFQGTVKELFYAKIRDAFNHPQFQTDVVR 113
+P VSYKPQ ++ K G+V++L + KIRDA+ HPQF TDV++
Sbjct 402 GEVPVLNVSYKPQKISPKSTGSVRQLLHEKIRDAYTHPQFVTDVMK 447
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 12/27 (44%), Positives = 20/27 (74%), Gaps = 0/27 (0%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPD 65
E++ ++G NG GKST +++LAG P+
Sbjct 104 EVLGLVGTNGIGKSTALKILAGKQKPN 130
> hsa:6059 ABCE1, ABC38, OABP, RLI, RNASEL1, RNASELI, RNS4I; ATP-binding
cassette, sub-family E (OABP), member 1; K06174 ATP-binding
cassette, sub-family E, member 1
Length=599
Score = 130 bits (327), Expect = 9e-31, Method: Compositional matrix adjust.
Identities = 57/106 (53%), Positives = 79/106 (74%), Gaps = 0/106 (0%)
Query 8 VQRLHFYEYPALTKTLGTFKLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNS 67
V+++ Y+YP + K +G F+L + G F+DSEI+VMLG+NGTGK+TFIRMLAG L PD
Sbjct 342 VKKMCMYKYPGMKKKMGEFELAIVAGEFTDSEIMVMLGENGTGKTTFIRMLAGRLKPDEG 401
Query 68 EALPSFAVSYKPQIVTAKFQGTVKELFYAKIRDAFNHPQFQTDVVR 113
+P VSYKPQ ++ K G+V++L + KIRDA+ HPQF TDV++
Sbjct 402 GEVPVLNVSYKPQKISPKSTGSVRQLLHEKIRDAYTHPQFVTDVMK 447
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 12/27 (44%), Positives = 20/27 (74%), Gaps = 0/27 (0%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPD 65
E++ ++G NG GKST +++LAG P+
Sbjct 104 EVLGLVGTNGIGKSTALKILAGKQKPN 130
> cpv:cgd1_980 RNase L inhibitor-like protein
Length=618
Score = 130 bits (326), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 62/126 (49%), Positives = 89/126 (70%), Gaps = 13/126 (10%)
Query 1 GLGFRV------WVQRLHFYEYPALTKTLGTFKLNVEKGSFSDSEIVVMLGQNGTGKSTF 54
GL F++ +QR +F +YP TKT+G+FKL+ E G F +SEI+VMLGQNG GK+TF
Sbjct 341 GLNFKIVDQDEIAMQRSNFVKYPGFTKTMGSFKLSAEAGDFGNSEIIVMLGQNGCGKTTF 400
Query 55 IRMLAGLLSPDNSEA-------LPSFAVSYKPQIVTAKFQGTVKELFYAKIRDAFNHPQF 107
I++LAG+ PD+S+ +P F VSYKPQ ++ KF+G+V++LF KIRD+F QF
Sbjct 401 IKILAGVSKPDDSDVIDKDHYQMPEFNVSYKPQTISPKFEGSVRDLFLMKIRDSFMDVQF 460
Query 108 QTDVVR 113
++VV+
Sbjct 461 TSEVVK 466
Score = 32.0 bits (71), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 17/63 (26%), Positives = 38/63 (60%), Gaps = 4/63 (6%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPDNSEALPSFAVSYKPQIVTAKFQGTVKELFYAKI 98
+++ ++G NG GKST +++L+G L P+ L + + + V A F+G+ + ++ K+
Sbjct 117 QVLGLVGTNGIGKSTALQILSGKLRPN----LGDYTKELEWKEVIAYFRGSELQTYFNKM 172
Query 99 RDA 101
++
Sbjct 173 QNG 175
> dre:406324 abce1, MGC56045, wu:fb34c09, wu:fe47b01, wu:fi09g07,
zgc:111906, zgc:56045; ATP-binding cassette, sub-family
E (OABP), member 1; K06174 ATP-binding cassette, sub-family
E, member 1
Length=599
Score = 127 bits (318), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 57/106 (53%), Positives = 79/106 (74%), Gaps = 0/106 (0%)
Query 8 VQRLHFYEYPALTKTLGTFKLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNS 67
V++ Y+YP + K++G F L + +G F+DSEI+VMLG+NGTGK+TFIRMLAG L PD
Sbjct 342 VKKPCRYQYPNMKKSMGEFTLTITEGEFTDSEIMVMLGENGTGKTTFIRMLAGGLKPDGG 401
Query 68 EALPSFAVSYKPQIVTAKFQGTVKELFYAKIRDAFNHPQFQTDVVR 113
+P VSYKPQ ++ KF+G+V+ L + KIRDA+ PQF TDV++
Sbjct 402 GDVPILNVSYKPQKISPKFKGSVRALLHDKIRDAYTRPQFVTDVMK 447
Score = 33.5 bits (75), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 39/73 (53%), Gaps = 8/73 (10%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPDNSEALPSFAVSYKPQIVTAKFQGTVKELFYAKI 98
E++ ++G NG GKST +++LAG P+ L F Q + A F+G+ + ++ KI
Sbjct 104 EVLGLVGTNGIGKSTALKILAGKQKPN----LGRFDAPPDWQEILAYFRGSELQNYFTKI 159
Query 99 RD----AFNHPQF 107
+ A PQ+
Sbjct 160 LEDDLKAIVKPQY 172
> cel:Y39E4B.1 abce-1; ABC transporter, class E family member
(abce-1); K06174 ATP-binding cassette, sub-family E, member
1
Length=610
Score = 122 bits (305), Expect = 4e-28, Method: Composition-based stats.
Identities = 54/106 (50%), Positives = 80/106 (75%), Gaps = 1/106 (0%)
Query 8 VQRLHFYEYPALTKTLGTFKLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSP-DN 66
++R +YP+++KTLG F L+VE G FSDSEI+VMLG+NGTGK+T I+M+AG L P D
Sbjct 351 IKRTGNIKYPSMSKTLGNFHLDVEAGDFSDSEIIVMLGENGTGKTTMIKMMAGSLKPEDE 410
Query 67 SEALPSFAVSYKPQIVTAKFQGTVKELFYAKIRDAFNHPQFQTDVV 112
+ LP ++SYKPQ ++ K + TV+ + + KI++ + HPQF+TDV+
Sbjct 411 NTELPHVSISYKPQKISPKSETTVRFMLHDKIQNMYEHPQFKTDVM 456
Score = 32.3 bits (72), Expect = 0.35, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 34/60 (56%), Gaps = 4/60 (6%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPDNSEALPSFAVSYKPQIVTAKFQGTVKELFYAKI 98
E++ ++G NG GKST +++LAG P+ L +F + + F+G+ + ++ +I
Sbjct 115 EVLGLVGTNGIGKSTALKILAGKQKPN----LGNFQKEQEWTTIINHFRGSELQNYFTRI 170
> ath:AT3G13640 ATRLI1; ATRLI1; transporter
Length=603
Score = 120 bits (301), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 57/102 (55%), Positives = 74/102 (72%), Gaps = 2/102 (1%)
Query 14 YEYPALTKTLGTFKLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDN--SEALP 71
Y+YP +TK LG FKL V +G F+DS+I+VMLG+NGTGK+TFIRMLAG + +P
Sbjct 350 YKYPNMTKQLGDFKLEVMEGEFTDSQIIVMLGENGTGKTTFIRMLAGAFPREEGVQSEIP 409
Query 72 SFAVSYKPQIVTAKFQGTVKELFYAKIRDAFNHPQFQTDVVR 113
F VSYKPQ +K + TV++L + KIRDA HPQF +DV+R
Sbjct 410 EFNVSYKPQGNDSKRECTVRQLLHDKIRDACAHPQFMSDVIR 451
Score = 31.2 bits (69), Expect = 0.80, Method: Compositional matrix adjust.
Identities = 12/27 (44%), Positives = 21/27 (77%), Gaps = 0/27 (0%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPD 65
+++ ++G NG GKST +++LAG L P+
Sbjct 104 QVLGLVGTNGIGKSTALKILAGKLKPN 130
> ath:AT4G30300 ATNAP15; transporter
Length=181
Score = 88.6 bits (218), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 47/101 (46%), Positives = 69/101 (68%), Gaps = 6/101 (5%)
Query 19 LTKTLGTFKLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNSEA-----LPSF 73
+T T G FKL + +G F+DS+I+VMLG+NGTGK+TFI+MLAG + E +P F
Sbjct 1 MTITRGDFKLRLNQGEFTDSQIIVMLGENGTGKTTFIKMLAGSKLDIDEEGSQVHEIPQF 60
Query 74 AVSYKPQ-IVTAKFQGTVKELFYAKIRDAFNHPQFQTDVVR 113
+VSYK Q + KF+ TV++L + KI +A+ QF +DV++
Sbjct 61 SVSYKNQHMSNRKFEITVRDLIHRKIPNAYAEHQFVSDVMK 101
> ath:AT1G63270 ATNAP10; transporter
Length=229
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 32/69 (46%), Gaps = 8/69 (11%)
Query 34 SFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNSEAL--------PSFAVSYKPQIVTAK 85
S D +V+ G NG+GKSTF+RMLAG P E L YK Q+
Sbjct 32 SLHDGGALVLTGTNGSGKSTFLRMLAGFSKPSAGEILWNGHDITQSGIFQQYKLQLNWIS 91
Query 86 FQGTVKELF 94
+ +KE F
Sbjct 92 LKDAIKERF 100
> ath:AT2G41700 ATPase, coupled to transmembrane movement of substances
/ amino acid transmembrane transporter
Length=1846
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 18/34 (52%), Positives = 25/34 (73%), Gaps = 0/34 (0%)
Query 37 DSEIVVMLGQNGTGKSTFIRMLAGLLSPDNSEAL 70
+++I+ +LG NG GKST I ML GLL P + +AL
Sbjct 507 ENQILSLLGHNGAGKSTTISMLVGLLPPTSGDAL 540
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 41/94 (43%), Gaps = 16/94 (17%)
Query 27 KLNVEKGSFS--DSEIVVMLGQNGTGKSTFIRMLAGLLSPDNSEAL---------PSFA- 74
K+ V+ +FS E LG NG GK+T + ML+G +P + A P
Sbjct 1429 KVAVQSLTFSVQAGECFGFLGTNGAGKTTTLSMLSGEETPTSGTAFIFGKDIVASPKAIR 1488
Query 75 --VSYKPQIVTAKFQGTVKEL--FYAKIRDAFNH 104
+ Y PQ TVKE YA+I+ +H
Sbjct 1489 QHIGYCPQFDALFEYLTVKEHLELYARIKGVVDH 1522
> ath:AT5G61700 ATATH16; ATPase, coupled to transmembrane movement
of substances / transporter
Length=888
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 36/81 (44%), Gaps = 14/81 (17%)
Query 34 SFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNSEAL--------------PSFAVSYKP 79
S S E MLG NG GK++FI M+ GLL P + AL S V +
Sbjct 593 SVSSGECFGMLGPNGAGKTSFISMMTGLLKPSSGTALVQGLDICKDMNKVYTSMGVCPQH 652
Query 80 QIVTAKFQGTVKELFYAKIRD 100
++ G LFY ++++
Sbjct 653 DLLWETLTGREHLLFYGRLKN 673
> dre:100332595 sugar abc transporter, putative-like; K02056 simple
sugar transport system ATP-binding protein [EC:3.6.3.17]
Length=518
Score = 40.4 bits (93), Expect = 0.002, Method: Composition-based stats.
Identities = 21/50 (42%), Positives = 31/50 (62%), Gaps = 7/50 (14%)
Query 19 LTKTLGTFK----LNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSP 64
LTK G+F +++E EI +LG+NG GKST ++ML G+L+P
Sbjct 13 LTKLFGSFAACNGIDIE---IQPGEIHALLGENGAGKSTLVKMLFGVLAP 59
> eco:b2547 yphE, ECK2544, JW2531; fused predicted sugar transporter
subunits of ABC superfamily: ATP-binding components;
K02056 simple sugar transport system ATP-binding protein [EC:3.6.3.17]
Length=503
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 24 GTFKLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNSE 68
G L+ + + E+ +LG+NG GKST IRML G PD+ +
Sbjct 22 GVVALDNVNFTLNKGEVRALLGKNGAGKSTLIRMLTGSERPDSGD 66
> eco:b4106 phnC, ECK4099, JW4067; phosphonate/organophosphate
ester transporter subunit; K02041 phosphonate transport system
ATP-binding protein
Length=262
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 29/47 (61%), Gaps = 5/47 (10%)
Query 21 KTLGTFKLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNS 67
+ L LN+ G E+V +LG +G+GKST +R L+GL++ D S
Sbjct 18 QALHAVDLNIHHG-----EMVALLGPSGSGKSTLLRHLSGLITGDKS 59
> eco:b3352 yheS, ECK3340, JW3315; fused predicted transporter
subunits of ABC superfamily: ATP-binding components; K06158
ATP-binding cassette, sub-family F, member 3
Length=637
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 21/46 (45%), Positives = 30/46 (65%), Gaps = 5/46 (10%)
Query 23 LGTFKLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNSE 68
L + KLN+ GS + +LG+NG GKST I++LAG L+P + E
Sbjct 328 LDSIKLNLVPGSR-----IGLLGRNGAGKSTLIKLLAGELAPVSGE 368
> ath:AT1G65410 ATNAP11 (ARABIDOPSIS THALIANA NON-INTRINSIC ABC
PROTEIN 11); transporter; K02065 putative ABC transport system
ATP-binding protein
Length=345
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 16/30 (53%), Positives = 24/30 (80%), Gaps = 0/30 (0%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPDNSE 68
E V ++G +GTGKST ++++AGLL+PD E
Sbjct 111 EAVGVIGPSGTGKSTILKIMAGLLAPDKGE 140
> eco:b1290 sapF, ECK1285, JW1283; antimicrobial peptide transport
ABC system ATP-binding protein; K02032 peptide/nickel transport
system ATP-binding protein
Length=268
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 26/37 (70%), Gaps = 0/37 (0%)
Query 34 SFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNSEAL 70
+ + + + ++G+NG+GKST +MLAG++ P + E L
Sbjct 36 TLREGQTLAIIGENGSGKSTLAKMLAGMIEPTSGELL 72
> ath:AT5G61740 ATATH14; ATPase, coupled to transmembrane movement
of substances / transporter
Length=848
Score = 38.5 bits (88), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 34/76 (44%), Gaps = 14/76 (18%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPDNSEAL--------------PSFAVSYKPQIVTA 84
E MLG NG GK++FI M+ GLL P + AL S V + ++
Sbjct 558 ECFGMLGPNGAGKTSFINMMTGLLKPTSGTALVQGLDICKDMNKVYTSMGVCPQHDLLWG 617
Query 85 KFQGTVKELFYAKIRD 100
G LFY ++++
Sbjct 618 TLTGREHLLFYGRLKN 633
> ath:AT3G47780 ATATH6; ATATH6; ATPase, coupled to transmembrane
movement of substances / transporter
Length=935
Score = 37.7 bits (86), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 34/76 (44%), Gaps = 14/76 (18%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPDNSEAL--------------PSFAVSYKPQIVTA 84
E MLG NG GK++FI M+ GLL P + AL S V + ++
Sbjct 645 ECFGMLGPNGAGKTSFINMMTGLLKPTSGTALVQGLDICNDMDRVYTSMGVCPQHDLLWE 704
Query 85 KFQGTVKELFYAKIRD 100
G LFY ++++
Sbjct 705 TLTGREHLLFYGRLKN 720
> eco:b3450 ugpC, ECK3434, JW3415; glycerol-3-phosphate transporter
subunit; K05816 sn-glycerol 3-phosphate transport system
ATP-binding protein [EC:3.6.3.20]
Length=356
Score = 37.4 bits (85), Expect = 0.011, Method: Composition-based stats.
Identities = 15/26 (57%), Positives = 21/26 (80%), Gaps = 0/26 (0%)
Query 36 SDSEIVVMLGQNGTGKSTFIRMLAGL 61
+D E +VM+G +G GKST +RM+AGL
Sbjct 28 ADGEFIVMVGPSGCGKSTLLRMVAGL 53
> eco:b0794 ybhF, ECK0783, JW5104; fused predicted transporter
subunits of ABC superfamily: ATP-binding components; K01990
ABC-2 type transport system ATP-binding protein
Length=578
Score = 37.4 bits (85), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 40 IVVMLGQNGTGKSTFIRMLAGLLSPDNSEA 69
+ ++G +G GK+T +RMLAGLL PD+ A
Sbjct 35 VTGLVGPDGAGKTTLMRMLAGLLKPDSGSA 64
Score = 37.4 bits (85), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 23/57 (40%), Positives = 29/57 (50%), Gaps = 1/57 (1%)
Query 15 EYPALTKTLGTFKLNVEKG-SFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNSEAL 70
E LTK G F + EI +LG NG GKST +M+ GLL P + +AL
Sbjct 331 EAKELTKKFGDFAATDHVNFAVKRGEIFGLLGPNGAGKSTTFKMMCGLLVPTSGQAL 387
> pfa:PFC0875w ABC transporter, putative
Length=3133
Score = 37.0 bits (84), Expect = 0.014, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 29/45 (64%), Gaps = 1/45 (2%)
Query 34 SFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNSEALPSFAVSYK 78
+ + I V+LG+NG+GKST I ++ ++S D+ E + F SYK
Sbjct 945 TLRSNRIFVLLGENGSGKSTLINIITKMISKDSGE-INFFKNSYK 988
> eco:b4391 yjjK, ECK4383, JW4354; fused predicted transporter
subunits of ABC superfamily: ATP-binding components
Length=555
Score = 37.0 bits (84), Expect = 0.016, Method: Composition-based stats.
Identities = 23/72 (31%), Positives = 34/72 (47%), Gaps = 2/72 (2%)
Query 34 SFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNSEALPS--FAVSYKPQIVTAKFQGTVK 91
SF + +LG NG GKST +R++AG+ EA P + Y PQ + TV+
Sbjct 28 SFFPGAKIGVLGLNGAGKSTLLRIMAGIDKDIEGEARPQPDIKIGYLPQEPQLNPEHTVR 87
Query 92 ELFYAKIRDAFN 103
E + + N
Sbjct 88 ESIEEAVSEVVN 99
Score = 35.4 bits (80), Expect = 0.043, Method: Composition-based stats.
Identities = 22/54 (40%), Positives = 31/54 (57%), Gaps = 3/54 (5%)
Query 15 EYPALTKTLGTFKLNVEKGSFS--DSEIVVMLGQNGTGKSTFIRMLAGLLSPDN 66
E L K+ G +L ++ SFS IV ++G NG GKST RM++G PD+
Sbjct 325 EVSNLRKSYGD-RLLIDDLSFSIPKGAIVGIIGPNGAGKSTLFRMISGQEQPDS 377
> eco:b1858 znuC, ECK1859, JW1847, yebM; zinc transporter subunit:
ATP-binding component of ABC superfamily; K09817 zinc transport
system ATP-binding protein [EC:3.6.3.-]
Length=251
Score = 37.0 bits (84), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 27/44 (61%), Gaps = 2/44 (4%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPDNS--EALPSFAVSYKPQ 80
+I+ +LG NG GKST +R++ GL++PD + + Y PQ
Sbjct 31 KILTLLGPNGAGKSTLVRVVLGLVTPDEGVIKRNGKLRIGYVPQ 74
> sce:YPL226W NEW1; ATP binding cassette protein that cosediments
with polysomes and is required for biogenesis of the small
ribosomal subunit; Asn/Gln-rich rich region supports [NU+]
prion formation and susceptibility to [PSI+] prion induction;
K03235 elongation factor 3
Length=1196
Score = 36.6 bits (83), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 21/49 (42%), Positives = 26/49 (53%), Gaps = 2/49 (4%)
Query 34 SFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNS--EALPSFAVSYKPQ 80
S S S V LG NG GKST I++L G L P+ E P+ + Y Q
Sbjct 835 SLSLSSRVACLGPNGAGKSTLIKLLTGELVPNEGKVEKHPNLRIGYIAQ 883
> mmu:320631 Abca15, 4930500I12Rik; ATP-binding cassette, sub-family
A (ABC1), member 15
Length=1668
Score = 36.6 bits (83), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 37 DSEIVVMLGQNGTGKSTFIRMLAGLLSPDNSEA 69
+ ++ V+LG NG GKST + +L+GL P + EA
Sbjct 550 EGQVTVLLGHNGAGKSTTLSILSGLYPPTSGEA 582
> ath:AT3G55100 ABC transporter family protein
Length=662
Score = 36.6 bits (83), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 34/70 (48%), Gaps = 12/70 (17%)
Query 11 LHFYEYPALTKTLGTFKLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPD----- 65
L F PA KTL LN G + EI+ +LG +G GKST I LAG ++
Sbjct 39 LRFGHSPAKIKTL----LNGITGEAKEGEILAILGASGAGKSTLIDALAGQIAEGSLKGT 94
Query 66 ---NSEALPS 72
N EAL S
Sbjct 95 VTLNGEALQS 104
> ath:AT3G47750 ABCA4; ABCA4 (ATP BINDING CASSETTE SUBFAMILY A4);
ATPase, coupled to transmembrane movement of substances
/ transporter
Length=946
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 16/32 (50%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPDNSEAL 70
E MLG NG GK++FI M+ GL+ P + AL
Sbjct 656 ECFGMLGPNGAGKTSFINMMTGLVKPTSGTAL 687
> ath:AT3G47770 ATATH5; ATATH5; ATPase, coupled to transmembrane
movement of substances / transporter
Length=900
Score = 36.2 bits (82), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 22/80 (27%), Positives = 34/80 (42%), Gaps = 14/80 (17%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSP--------------DNSEALPSFAVSYKPQIVTA 84
E MLG NG GK++FI M+ GL+ P D + S V + ++
Sbjct 616 ECFGMLGPNGAGKTSFINMMTGLVKPSSGSAFVQGLDICKDMDKVYISMGVCPQHDLLWE 675
Query 85 KFQGTVKELFYAKIRDAFNH 104
G LFY ++++ H
Sbjct 676 TLTGKEHLLFYGRLKNLKGH 695
> dre:564344 ABC Transporter family member (abt-4)-like; K05643
ATP-binding cassette, subfamily A (ABC1), member 3
Length=1343
Score = 36.2 bits (82), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 37 DSEIVVMLGQNGTGKSTFIRMLAGLLSPDNSEA 69
+ +I V+LG NG GK+T + ML GL P + A
Sbjct 194 EGQITVLLGHNGAGKTTTLSMLTGLFPPSSGRA 226
> eco:b2129 yehX, ECK2122, JW2117; predicted transporter subunit:
ATP-binding component of ABC superfamily; K05847 osmoprotectant
transport system ATP-binding protein
Length=308
Score = 35.8 bits (81), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 30/48 (62%), Gaps = 5/48 (10%)
Query 21 KTLGTFKLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNSE 68
K + LN ++GSFS V++G +G+GKST ++M+ L+ D+ E
Sbjct 15 KAVNDLNLNFQEGSFS-----VLIGTSGSGKSTTLKMINRLVEHDSGE 57
> mmu:381072 Abca17; ATP-binding cassette, sub-family A (ABC1),
member 17
Length=1733
Score = 35.8 bits (81), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 33/64 (51%), Gaps = 7/64 (10%)
Query 8 VQRLH--FYEYPALTKTLGTFKLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPD 65
+Q L+ FY + + +N+ KG ++ V+LG NG GK+T +L GL++P
Sbjct 521 IQHLYKVFYSGRSKRTAIRDLSMNLYKG-----QVTVLLGHNGAGKTTVCSVLTGLITPS 575
Query 66 NSEA 69
A
Sbjct 576 KGHA 579
> hsa:21 ABCA3, ABC-C, ABC3, EST111653, LBM180, MGC166979, MGC72201,
SMDP3; ATP-binding cassette, sub-family A (ABC1), member
3; K05643 ATP-binding cassette, subfamily A (ABC1), member
3
Length=1704
Score = 35.8 bits (81), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 37 DSEIVVMLGQNGTGKSTFIRMLAGLLSPDNSEA 69
+ +I V+LG NG GK+T + ML GL P + A
Sbjct 558 EGQITVLLGHNGAGKTTTLSMLTGLFPPTSGRA 590
> eco:b0066 thiQ, ECK0067, JW0065, sfuC, yabJ; thiamin transporter
subunit; K02062 thiamine transport system ATP-binding protein
Length=232
Score = 35.8 bits (81), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 18/39 (46%), Positives = 25/39 (64%), Gaps = 5/39 (12%)
Query 26 FKLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSP 64
F L VE+G E V +LG +G GKST + ++AG L+P
Sbjct 18 FSLTVERG-----EQVAILGPSGAGKSTLLNLIAGFLTP 51
> ath:AT3G47790 ATATH7; ATATH7; ATPase, coupled to transmembrane
movement of substances / transporter
Length=901
Score = 35.8 bits (81), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 20/76 (26%), Positives = 34/76 (44%), Gaps = 14/76 (18%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPDNSEAL--------------PSFAVSYKPQIVTA 84
E MLG NG GK++FI M+ G++ P + A + V + ++
Sbjct 618 ECFGMLGPNGAGKTSFINMMTGIIKPSSGTAFVQGLDILTDMDRIYTTIGVCPQHDLLWE 677
Query 85 KFQGTVKELFYAKIRD 100
K G LFY ++++
Sbjct 678 KLSGREHLLFYGRLKN 693
> ath:AT5G09930 ATGCN2; ATGCN2; transporter
Length=678
Score = 35.8 bits (81), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 33/67 (49%), Gaps = 3/67 (4%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPDNSE---ALPSFAVSYKPQIVTAKFQGTVKELFY 95
E V ++G NG GK+T +R++ G PD+ A P+ V++ Q TVKE F
Sbjct 110 EKVGLIGVNGAGKTTQLRIITGQEEPDSGNVIWAKPNLKVAFLSQEFEVSMGKTVKEEFM 169
Query 96 AKIRDAF 102
++
Sbjct 170 CTFKEEM 176
Score = 32.7 bits (73), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 25/43 (58%), Gaps = 5/43 (11%)
Query 28 LNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNSEAL 70
L +E+G E V ++G NG GKST ++++ GL P E +
Sbjct 431 LAIERG-----EKVAIIGPNGCGKSTLLKLIMGLEKPMRGEVI 468
> eco:b2211 yojI, ECK2203, JW2199, yojJ; Microcin J25 efflux pump,
TolC-dependent; fused ABC transporter permease and ATP-binding
components; K06159 putative ATP-binding cassette transporter
Length=547
Score = 35.8 bits (81), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 29/49 (59%), Gaps = 5/49 (10%)
Query 22 TLGTFKLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNSEAL 70
++G L +++G E++ ++G NG+GKST +L GL P + E L
Sbjct 338 SVGPINLTIKRG-----ELLFLIGGNGSGKSTLAMLLTGLYQPQSGEIL 381
> eco:b0855 potG, ECK0846, JW5818; putrescine transporter subunit:
ATP-binding component of ABC superfamily; K11076 putrescine
transport system ATP-binding protein
Length=377
Score = 35.4 bits (80), Expect = 0.041, Method: Composition-based stats.
Identities = 15/32 (46%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPDNSEAL 70
EI +LG +G GKST +RMLAG P + +
Sbjct 46 EIFALLGASGCGKSTLLRMLAGFEQPSAGQIM 77
> ath:AT3G47760 ATATH4; ATATH4; ATPase, coupled to transmembrane
movement of substances / transporter
Length=872
Score = 35.4 bits (80), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 21/76 (27%), Positives = 33/76 (43%), Gaps = 14/76 (18%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPDNSEAL--------------PSFAVSYKPQIVTA 84
E MLG NG GK++FI M+ GL+ P + A S V + ++
Sbjct 582 ECFGMLGPNGAGKTSFINMMTGLMKPTSGAAFVHGLDICKDMDIVYTSIGVCPQHDLLWE 641
Query 85 KFQGTVKELFYAKIRD 100
G LFY ++++
Sbjct 642 TLTGREHLLFYGRLKN 657
> eco:b3480 nikE, ECK3464, hydC, hydD, JW3445; nickel transporter
subunit; K02032 peptide/nickel transport system ATP-binding
protein
Length=268
Score = 35.4 bits (80), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 15/31 (48%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 34 SFSDSEIVVMLGQNGTGKSTFIRMLAGLLSP 64
+ E V +LG++G GKST R+L GL SP
Sbjct 34 TLKSGETVALLGRSGCGKSTLARLLVGLESP 64
> mmu:11304 Abca4, AW050280, Abc10, Abcr, D430003I15Rik, RmP;
ATP-binding cassette, sub-family A (ABC1), member 4; K05644
ATP-binding cassette, subfamily A (ABC1), member 4
Length=2310
Score = 35.4 bits (80), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 27/44 (61%), Gaps = 3/44 (6%)
Query 27 KLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNSEAL 70
+LN+ +F +++I LG NG GK+T + +L GLL P + L
Sbjct 948 RLNI---TFYENQITAFLGHNGAGKTTTLSILTGLLPPTSGTVL 988
> hsa:24 ABCA4, ABC10, ABCR, ARMD2, CORD3, DKFZp781N1972, FFM,
FLJ17534, RMP, RP19, STGD, STGD1; ATP-binding cassette, sub-family
A (ABC1), member 4; K05644 ATP-binding cassette, subfamily
A (ABC1), member 4
Length=2273
Score = 35.4 bits (80), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 27/44 (61%), Gaps = 3/44 (6%)
Query 27 KLNVEKGSFSDSEIVVMLGQNGTGKSTFIRMLAGLLSPDNSEAL 70
+LN+ +F +++I LG NG GK+T + +L GLL P + L
Sbjct 948 RLNI---TFYENQITAFLGHNGAGKTTTLSILTGLLPPTSGTVL 988
> mmu:268379 Abca13, 9830132L24, A930002G16Rik, AI956815; ATP-binding
cassette, sub-family A (ABC1), member 13; K05647 ATP
binding cassette, subfamily A (ABC1), member 13
Length=5034
Score = 35.4 bits (80), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 15/31 (48%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 34 SFSDSEIVVMLGQNGTGKSTFIRMLAGLLSP 64
+F +I +LG NG GK+T I ML GL P
Sbjct 3832 TFHRDQITALLGTNGAGKTTIISMLMGLFPP 3862
> ath:AT5G64840 ATGCN5; ATGCN5 (A. THALIANA GENERAL CONTROL NON-REPRESSIBLE
5); transporter
Length=692
Score = 35.4 bits (80), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 20/67 (29%), Positives = 34/67 (50%), Gaps = 3/67 (4%)
Query 39 EIVVMLGQNGTGKSTFIRMLAGLLSPDNS---EALPSFAVSYKPQIVTAKFQGTVKELFY 95
E V ++G NG GK+T +R++ G PD+ +A P+ V++ Q TV+E F
Sbjct 124 EKVGLVGVNGAGKTTQLRIITGQEEPDSGNVIKAKPNMKVAFLSQEFEVSMSKTVREEFM 183
Query 96 AKIRDAF 102
++
Sbjct 184 TAFKEEM 190
Lambda K H
0.322 0.137 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2052546600
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40