bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
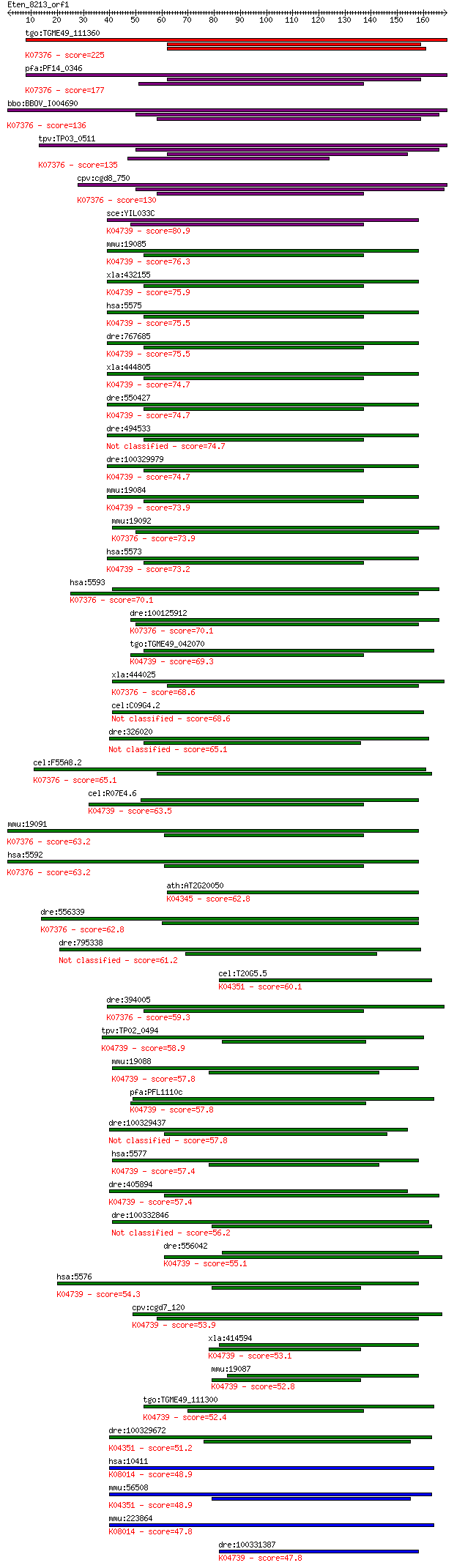
Query= Eten_8213_orf1
Length=168
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_111360 cGMP-dependent protein kinase, putative (EC:... 225 5e-59
pfa:PF14_0346 PKG; cGMP-dependent protein kinase (EC:2.7.11.12... 177 2e-44
bbo:BBOV_I004690 19.m02237; cGMP dependent protein kinase (EC:... 136 4e-32
tpv:TP03_0511 cGMP-dependent protein kinase (EC:2.7.1.37); K07... 135 7e-32
cpv:cgd8_750 cyclic nucleotide (cGMP)-dependent protein kinase... 130 3e-30
sce:YIL033C BCY1, SRA1; Bcy1p; K04739 cAMP-dependent protein k... 80.9 2e-15
mmu:19085 Prkar1b, AI385716, RIbeta; protein kinase, cAMP depe... 76.3 5e-14
xla:432155 prkar1a, pkr1; protein kinase, cAMP-dependent, regu... 75.9 6e-14
hsa:5575 PRKAR1B, PRKAR1; protein kinase, cAMP-dependent, regu... 75.5 7e-14
dre:767685 prkar1b, MGC153624, zgc:153624; protein kinase, cAM... 75.5 8e-14
xla:444805 prkar1b, MGC82149, prkar1; protein kinase, cAMP-dep... 74.7 1e-13
dre:550427 prkar1ab, zgc:112145; protein kinase, cAMP-dependen... 74.7 1e-13
dre:494533 prkar1aa, im:7047729, prkar1a, zgc:92515; protein k... 74.7 1e-13
dre:100329979 protein kinase, cAMP-dependent, regulatory, type... 74.7 1e-13
mmu:19084 Prkar1a, 1300018C22Rik, RIalpha, Tse-1, Tse1; protei... 73.9 2e-13
mmu:19092 Prkg2, AW212535, CGKII, MGC130517, Prkgr2, cGK-II; p... 73.9 3e-13
hsa:5573 PRKAR1A, CAR, CNC, CNC1, DKFZp779L0468, MGC17251, PKR... 73.2 4e-13
hsa:5593 PRKG2, PRKGR2, cGKII; protein kinase, cGMP-dependent,... 70.1 3e-12
dre:100125912 prkg2; protein kinase, cGMP-dependent, type II (... 70.1 4e-12
tgo:TGME49_042070 cAMP-dependent protein kinase regulatory sub... 69.3 6e-12
xla:444025 prkg2, MGC82580; protein kinase, cGMP-dependent, ty... 68.6 9e-12
cel:C09G4.2 hypothetical protein 68.6 9e-12
dre:326020 fd58b04, wu:fd58b04; si:dkey-121j17.5 65.1 1e-10
cel:F55A8.2 egl-4; EGg Laying defective family member (egl-4);... 65.1 1e-10
cel:R07E4.6 kin-2; protein KINase family member (kin-2); K0473... 63.5 3e-10
mmu:19091 Prkg1, AW125416, CGKI, MGC132849, Prkg1b, Prkgr1b; p... 63.2 4e-10
hsa:5592 PRKG1, CGKI, DKFZp686K042, FLJ36117, MGC71944, PGK, P... 63.2 4e-10
ath:AT2G20050 ATP binding / cAMP-dependent protein kinase regu... 62.8 5e-10
dre:556339 protein kinase, cGMP-dependent, type II-like; K0737... 62.8 6e-10
dre:795338 si:dkey-196h17.4 61.2 2e-09
cel:T20G5.5 epac-1; Exchange Protein Activated by Cyclic AMP f... 60.1 4e-09
dre:394005 prkg1a, MGC63955, prkg1, zgc:63955; protein kinase,... 59.3 6e-09
tpv:TP02_0494 cAMP-dependent protein kinase regulatory subunit... 58.9 9e-09
mmu:19088 Prkar2b, AI451071, AW061005, Pkarb2, RII(beta); prot... 57.8 2e-08
pfa:PFL1110c PKAr; CAMP-dependent protein kinase regulatory su... 57.8 2e-08
dre:100329437 protein kinase, cAMP-dependent, regulatory, type... 57.8 2e-08
hsa:5577 PRKAR2B, PRKAR2, RII-BETA; protein kinase, cAMP-depen... 57.4 2e-08
dre:405894 prkar2ab, MGC85886, prkar2a, zgc:85886; protein kin... 57.4 2e-08
dre:100332846 acyl-CoA synthetase long-chain family member 6-like 56.2 6e-08
dre:556042 prkar2aa, MGC153742, wu:fb55f12, wu:fj29e12, wu:fj3... 55.1 1e-07
hsa:5576 PRKAR2A, MGC3606, PKR2, PRKAR2; protein kinase, cAMP-... 54.3 2e-07
cpv:cgd7_120 cAMP-dependent protein kinase regulatory subunit ... 53.9 3e-07
xla:414594 prkar2b, MGC83177; protein kinase, cAMP-dependent, ... 53.1 4e-07
mmu:19087 Prkar2a, 1110061A24Rik, AI317181, AI836829, RII(alph... 52.8 5e-07
tgo:TGME49_111300 cAMP-dependent protein kinase regulatory sub... 52.4 7e-07
dre:100329672 cAMP-regulated guanine nucleotide exchange facto... 51.2 1e-06
hsa:10411 RAPGEF3, CAMP-GEFI, EPAC, EPAC1, HSU79275, MGC21410,... 48.9 8e-06
mmu:56508 Rapgef4, 1300003D15Rik, 5730402K07Rik, 6330581N18Rik... 48.9 9e-06
mmu:223864 Rapgef3, 2310016P22Rik, 9330170P05Rik, Epac, Epac1,... 47.8 2e-05
dre:100331387 cAMP-dependent protein kinase, regulatory subuni... 47.8 2e-05
> tgo:TGME49_111360 cGMP-dependent protein kinase, putative (EC:2.7.11.12);
K07376 protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=994
Score = 225 bits (574), Expect = 5e-59, Method: Compositional matrix adjust.
Identities = 108/162 (66%), Positives = 132/162 (81%), Gaps = 1/162 (0%)
Query 8 KHGGERK-VQKAIKQQEDTQAEDARLLGHLEKREKTPSDLSLIRDSLSTNLVCSSLNDAE 66
K GG+RK QKAI +Q+D+ E+ +L HL REKTP+D +LI+DSL NLVCSSLN+ E
Sbjct 138 KPGGDRKPAQKAILKQDDSHTEEEKLNAHLAYREKTPADFALIQDSLKANLVCSSLNEGE 197
Query 67 VEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKVVNKILTGQAFGEISL 126
++ALA A++FFTFKKGDVVTKQGE GSYFFI+HSG F+V+VNDK VN + G+AFGEI+L
Sbjct 198 IDALAVAMQFFTFKKGDVVTKQGEPGSYFFIIHSGTFDVLVNDKRVNAMDKGKAFGEIAL 257
Query 127 IHNSARTATIKTLSEDAALWGVQRQVFRETLKQLSSRNFAEN 168
IHN+ R+AT+ S + ALWGVQR FRETLKQLSSRNFAEN
Sbjct 258 IHNTERSATVVASSTEGALWGVQRHTFRETLKQLSSRNFAEN 299
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 31/97 (31%), Positives = 55/97 (56%), Gaps = 0/97 (0%)
Query 62 LNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKVVNKILTGQAF 121
L+D ++ L A + + G+ + K+GE G+ FFI+ +GE ++ N+K + + F
Sbjct 565 LSDHQMTMLIKAFKTVRYMSGEYIIKEGERGTRFFIIKAGEVAILKNNKRLRTLGRHDYF 624
Query 122 GEISLIHNSARTATIKTLSEDAALWGVQRQVFRETLK 158
GE +L+++ RTA++ S LW V + VF E +K
Sbjct 625 GERALLYDEPRTASVCANSAGVDLWVVDKSVFNEIIK 661
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 28/99 (28%), Positives = 51/99 (51%), Gaps = 1/99 (1%)
Query 62 LNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKVVNKILTGQAF 121
L +A+ + NA+ FK G + K+G++G +I+ SG+ +V + + + + G F
Sbjct 312 LTEAQKNVITNALVVENFKPGQPIVKEGDAGDVLYILKSGKAKVSIGGREIRMLRKGDYF 371
Query 122 GEISLIHNSARTATIKTLSEDAALWGVQRQVFRETLKQL 160
GE +L++ R+ATI T E + R++ L L
Sbjct 372 GERALLYKEPRSATI-TAEEFTVCVSIGRELLDRVLGNL 409
> pfa:PF14_0346 PKG; cGMP-dependent protein kinase (EC:2.7.11.12);
K07376 protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=853
Score = 177 bits (449), Expect = 2e-44, Method: Composition-based stats.
Identities = 88/161 (54%), Positives = 112/161 (69%), Gaps = 1/161 (0%)
Query 8 KHGGERKVQKAIKQQEDTQAEDARLLGHLEKREKTPSDLSLIRDSLSTNLVCSSLNDAEV 67
K G ER +KAI +D ED+ + HLE REK D+ +I+ SL NLVCS+LND E+
Sbjct 8 KKGNERNKKKAIFSNDDFTGEDSLMEDHLELREKLSEDIDMIKTSLKNNLVCSTLNDNEI 67
Query 68 EALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKVVNKILTGQAFGEISLI 127
L+N ++FF FK G++V KQGE GSYFFI++SG+F+V VNDK V + G +FGE +LI
Sbjct 68 LTLSNYMQFFVFKSGNLVIKQGEKGSYFFIINSGKFDVYVNDKKVKTMGKGSSFGEAALI 127
Query 128 HNSARTATIKTLSEDAALWGVQRQVFRETLKQLSSRNFAEN 168
HN+ R+ATI D LWGVQR FR TLKQLS+RNF EN
Sbjct 128 HNTQRSATI-IAETDGTLWGVQRSTFRATLKQLSNRNFNEN 167
Score = 60.1 bits (144), Expect = 3e-09, Method: Composition-based stats.
Identities = 28/97 (28%), Positives = 54/97 (55%), Gaps = 0/97 (0%)
Query 62 LNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKVVNKILTGQAF 121
L D + L A +++GD + ++GE GS F+I+ +GE E++ N K + + F
Sbjct 422 LTDKQCNLLIEAFRTTRYEEGDYIIQEGEVGSRFYIIKNGEVEIVKNKKRLRTLGKNDYF 481
Query 122 GEISLIHNSARTATIKTLSEDAALWGVQRQVFRETLK 158
GE +L+++ RTA++ + + W V + VF + ++
Sbjct 482 GERALLYDEPRTASVISKVNNVECWFVDKSVFLQIIQ 518
Score = 53.5 bits (127), Expect = 3e-07, Method: Composition-based stats.
Identities = 29/86 (33%), Positives = 48/86 (55%), Gaps = 3/86 (3%)
Query 51 DSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDK 110
DS+S V L +A+ + NA FK G+ + KQG+ G +I+ G+ V +ND+
Sbjct 172 DSVS---VFDMLTEAQKNMITNACVIQNFKSGETIVKQGDYGDVLYILKEGKATVYINDE 228
Query 111 VVNKILTGQAFGEISLIHNSARTATI 136
+ + G FGE +L+++ R+ATI
Sbjct 229 EIRVLEKGSYFGERALLYDEPRSATI 254
> bbo:BBOV_I004690 19.m02237; cGMP dependent protein kinase (EC:2.7.11.1);
K07376 protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=887
Score = 136 bits (342), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 69/173 (39%), Positives = 110/173 (63%), Gaps = 5/173 (2%)
Query 1 STAAAPHKHGGERKVQ-KAIKQQEDTQAEDARLLG-HLEKREKTPSDLSLIRDSLSTNLV 58
+ A P+ + G + V+ + + + + ++ L + R+KT D++LIR +L+ NLV
Sbjct 22 TKAWKPNNYQGSKGVRSQPVNVAHNEECDEVSTLAKYATSRDKTDKDITLIRHALANNLV 81
Query 59 CSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKVVNKILTG 118
C SLN+ E++A ++ F G V +QG++G+YFFI+ G+F+V V+ + VN + G
Sbjct 82 CESLNEYEIDAFIGSMSSFELPAGAPVVRQGDNGTYFFIISEGDFDVYVDGEHVNSMTRG 141
Query 119 QAFGEISLIHNSARTATIKTLSED---AALWGVQRQVFRETLKQLSSRNFAEN 168
AFGEISLIHN+ R+AT+K ++ LWGV R VFR+TLK++S RN+ EN
Sbjct 142 TAFGEISLIHNTPRSATVKVKNKAGNCGKLWGVTRVVFRDTLKRISLRNYTEN 194
Score = 55.8 bits (133), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 31/116 (26%), Positives = 58/116 (50%), Gaps = 1/116 (0%)
Query 50 RDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVND 109
R+ + + +L + + NA+ F G+ + KQG+ G +++ G +V VND
Sbjct 195 REFIDCVTIFENLTEENKSCITNALVELRFSPGESIVKQGDPGDDLYLIKQGSADVYVND 254
Query 110 KVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETLKQLSSRNF 165
V I GQ FGE +++++ R+ATIK + + + R + + L L++ F
Sbjct 255 IRVRTITKGQYFGERAILYSEPRSATIKAIDYVICV-SITRDILQSVLGNLNNVLF 309
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 26/101 (25%), Positives = 55/101 (54%), Gaps = 0/101 (0%)
Query 58 VCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKVVNKILT 117
+ L++ +++ L A++ +K D + +GE G +I+ SGE +I N + +
Sbjct 450 ILRHLSEKQIDMLIKALKTLRYKLNDTIFNEGEIGDMLYIIKSGEVAIIKNGVKIRTLGK 509
Query 118 GQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETLK 158
FGE +L+++ R+A++ + + LW V++ VF E ++
Sbjct 510 HDYFGERALLYDEQRSASVVSNATYVDLWVVEKPVFMELME 550
> tpv:TP03_0511 cGMP-dependent protein kinase (EC:2.7.1.37); K07376
protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=892
Score = 135 bits (340), Expect = 7e-32, Method: Composition-based stats.
Identities = 70/167 (41%), Positives = 113/167 (67%), Gaps = 11/167 (6%)
Query 13 RKVQKAIKQQEDTQA-EDARLLGHLEKREKTPSDLSLIRDSLSTNLVCSSLNDAEVEALA 71
++ K IK+ ++++ ED ++ HL KREK+ SD S I+ SL+ N++ S+LND E+ A
Sbjct 34 KQRSKFIKKANESESYEDTEIVKHLVKREKSESDKSFIKKSLANNVIFSALNDLEMSAFV 93
Query 72 NAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKVVNKILTGQAFGEISLIHNSA 131
+++ ++ F G VT+QG +GSYFF+++ G F+V ++ K+VN + G AFGEISLI+++
Sbjct 94 DSMSYYVFSVGSKVTEQGTNGSYFFVINEGIFDVYIDGKLVNTMERGTAFGEISLINDTP 153
Query 132 RTATIKT--LSED--------AALWGVQRQVFRETLKQLSSRNFAEN 168
RTAT+K ++E+ +LWGV R VFRETLK +S +++N
Sbjct 154 RTATVKVRGVTENVKASSDTLGSLWGVNRTVFRETLKNISMEVYSQN 200
Score = 56.2 bits (134), Expect = 5e-08, Method: Composition-based stats.
Identities = 33/116 (28%), Positives = 59/116 (50%), Gaps = 1/116 (0%)
Query 50 RDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVND 109
R L + + L + + + NA F GD + QG+ G +I+ G+ +V++ND
Sbjct 201 RSFLDSVKIFEMLTENQKNMVTNAFVESKFVPGDRIVTQGDFGDVLYIIKEGKADVLIND 260
Query 110 KVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETLKQLSSRNF 165
+ V + GQ FGE +L+++ R+ATI +ED + R++ + L L + F
Sbjct 261 EKVRTLTNGQYFGERALLYDEPRSATI-VATEDTVCVSLGREILNKVLGNLDNVLF 315
Score = 50.1 bits (118), Expect = 4e-06, Method: Composition-based stats.
Identities = 26/92 (28%), Positives = 54/92 (58%), Gaps = 0/92 (0%)
Query 62 LNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKVVNKILTGQAF 121
L++A ++ L + +KKG+ + ++GE G F+I+ G+ ++I + + + F
Sbjct 463 LSEAHLDMLIKKFKSKRYKKGEYIIREGEVGDSFYIISKGDVDIIKSGTRLRTLGKNDYF 522
Query 122 GEISLIHNSARTATIKTLSEDAALWGVQRQVF 153
GE +L+++ RTA++ S ++ LW V++ VF
Sbjct 523 GERALLYDEPRTASVVCSSANSDLWVVEKSVF 554
Score = 30.8 bits (68), Expect = 2.4, Method: Composition-based stats.
Identities = 19/79 (24%), Positives = 36/79 (45%), Gaps = 2/79 (2%)
Query 47 SLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGES--GSYFFIVHSGEFE 104
+++ + L + V ++ L + FK GDV+ + E G FFI+ G +
Sbjct 317 NVMMEYLQNSTVFRQFTPDQLAKLIESASVRDFKPGDVILHREEKIKGIRFFILLEGSVQ 376
Query 105 VIVNDKVVNKILTGQAFGE 123
++ D + + G +FGE
Sbjct 377 ILYEDNEIGIMERGDSFGE 395
> cpv:cgd8_750 cyclic nucleotide (cGMP)-dependent protein kinase
with 3 cNMP binding domains and a Ser/Thr kinase domain ;
K07376 protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=965
Score = 130 bits (326), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 61/141 (43%), Positives = 96/141 (68%), Gaps = 1/141 (0%)
Query 28 EDARLLGHLEKREKTPSDLSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTK 87
E ++ HL+ REKT D+ I +L+ N+V +SLN++E+ L +++ ++ ++ G+VV +
Sbjct 79 EQNMVISHLKDREKTEEDIKTISKALAGNVVGASLNESEIATLVSSMHYYEYEVGEVVIE 138
Query 88 QGESGSYFFIVHSGEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWG 147
QG SG YFF++ +G F V +N VN + G AFGE++LIHN+ R+ATI + E LWG
Sbjct 139 QGASGFYFFVISTGSFGVEINGNRVNTMSEGTAFGELALIHNTPRSATILVI-EKGGLWG 197
Query 148 VQRQVFRETLKQLSSRNFAEN 168
+ R FR+TL+ +SSRN+ EN
Sbjct 198 LGRSTFRDTLRLISSRNYEEN 218
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 32/118 (27%), Positives = 63/118 (53%), Gaps = 0/118 (0%)
Query 50 RDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVND 109
RD++ + V +++ ++ L ++ F G+ + QG+ G+ FFI+ SGE V N+
Sbjct 491 RDAIKSCFVFQYVSEQQLSLLVKSLRLVKFTSGEKIVVQGDKGTAFFILQSGEVAVYRNN 550
Query 110 KVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETLKQLSSRNFAE 167
K + + FGE +L+++ R+ATI+ + + LW V ++ F + ++ R E
Sbjct 551 KFIRYLGKNDYFGERALLYDELRSATIEAATPEVHLWTVDKEAFLKIVEPPMRRYLDE 608
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 26/79 (32%), Positives = 43/79 (54%), Gaps = 0/79 (0%)
Query 58 VCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKVVNKILT 117
+ S L D + L+ A+ F K V+ ++ E G+ +++ SG V V DK + +
Sbjct 227 IFSGLTDKQKSLLSEALVREIFVKDQVIIREKEIGNVLYMIKSGIVGVFVEDKYIRSLNE 286
Query 118 GQAFGEISLIHNSARTATI 136
G AFGE SL+ + R+AT+
Sbjct 287 GDAFGERSLMFDEPRSATV 305
> sce:YIL033C BCY1, SRA1; Bcy1p; K04739 cAMP-dependent protein
kinase regulator
Length=416
Score = 80.9 bits (198), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 41/119 (34%), Positives = 64/119 (53%), Gaps = 1/119 (0%)
Query 39 REKTPSDLSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIV 98
+EK+ L + S+ N + + L+ + N +E + KG + KQG+ G YF++V
Sbjct 165 KEKSEQQLQRLEKSIRNNFLFNKLDSDSKRLVINCLEEKSVPKGATIIKQGDQGDYFYVV 224
Query 99 HSGEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETL 157
G + VND VN G +FGE++L++NS R AT+ S D LW + R FR+ L
Sbjct 225 EKGTVDFYVNDNKVNSSGPGSSFGELALMYNSPRAATVVATS-DCLLWALDRLTFRKIL 282
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 24/90 (26%), Positives = 50/90 (55%), Gaps = 1/90 (1%)
Query 48 LIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIV 107
+ D L + V SL + LA+A++ ++ G+ + ++G+ G F+++ G +V
Sbjct 292 MYDDLLKSMPVLKSLTTYDRAKLADALDTKIYQPGETIIREGDQGENFYLIEYGAVDVSK 351
Query 108 NDK-VVNKILTGQAFGEISLIHNSARTATI 136
+ V+NK+ FGE++L+++ R AT+
Sbjct 352 KGQGVINKLKDHDYFGEVALLNDLPRQATV 381
> mmu:19085 Prkar1b, AI385716, RIbeta; protein kinase, cAMP dependent
regulatory, type I beta (EC:2.7.11.1); K04739 cAMP-dependent
protein kinase regulator
Length=381
Score = 76.3 bits (186), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 41/122 (33%), Positives = 69/122 (56%), Gaps = 4/122 (3%)
Query 39 REKTPSD---LSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYF 95
R+ P D ++ + ++S N++ S L+D E + +A+ T G+ V +QG G F
Sbjct 115 RKVIPKDYKTMTALAKAISKNVLFSHLDDNERSDIFDAMFPVTHIGGETVIQQGNEGDNF 174
Query 96 FIVHSGEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRE 155
+++ GE +V VN + V I G +FGE++LI+ + R AT+K + D LWG+ R +R
Sbjct 175 YVIDQGEVDVYVNGEWVTNISEGGSFGELALIYGTPRAATVKAKT-DLKLWGIDRDSYRR 233
Query 156 TL 157
L
Sbjct 234 IL 235
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 46/90 (51%), Gaps = 6/90 (6%)
Query 53 LSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVI----VN 108
LS + SL E +A+A+E F+ G+ + QGE G F+I+ G V+ N
Sbjct 250 LSKVSILESLEKWERLTVADALEPVQFEDGEKIVVQGEPGDDFYIITEGTASVLQRRSPN 309
Query 109 DKV--VNKILTGQAFGEISLIHNSARTATI 136
++ V ++ FGEI+L+ N R AT+
Sbjct 310 EEYVEVGRLGPSDYFGEIALLLNRPRAATV 339
> xla:432155 prkar1a, pkr1; protein kinase, cAMP-dependent, regulatory,
type I, alpha (tissue specific extinguisher 1) (EC:2.7.11.1);
K04739 cAMP-dependent protein kinase regulator
Length=381
Score = 75.9 bits (185), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 38/122 (31%), Positives = 71/122 (58%), Gaps = 4/122 (3%)
Query 39 REKTPSD---LSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYF 95
R+ P D ++ + ++ N++ + L+D E + +A+ T+ G+ V +QG+ G F
Sbjct 115 RKVIPKDYKTMAALAKAIEKNVLFAHLDDTERSDIFDAMFSVTYISGETVIQQGDEGDNF 174
Query 96 FIVHSGEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRE 155
++V GE +V VN++ + I G +FGE++LI+ + R AT+K + + LWG+ R +R
Sbjct 175 YVVDQGEMDVYVNNEWMTSIGEGGSFGELALIYGTPRAATVKAKT-NVKLWGIDRDSYRR 233
Query 156 TL 157
L
Sbjct 234 IL 235
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 46/90 (51%), Gaps = 6/90 (6%)
Query 53 LSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIV----N 108
LS + SL+ E +A+A+E F+ G + QGE G FFI+ G V+ N
Sbjct 250 LSKVSILESLDKWERLTVADALEPVQFEDGQKIVVQGEPGDEFFIILEGSAAVLQRRSEN 309
Query 109 DKV--VNKILTGQAFGEISLIHNSARTATI 136
++ V ++ FGEI+L+ N R AT+
Sbjct 310 EEFVEVGRLGPSDYFGEIALLMNRPRAATV 339
> hsa:5575 PRKAR1B, PRKAR1; protein kinase, cAMP-dependent, regulatory,
type I, beta (EC:2.7.11.1); K04739 cAMP-dependent
protein kinase regulator
Length=381
Score = 75.5 bits (184), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 41/122 (33%), Positives = 69/122 (56%), Gaps = 4/122 (3%)
Query 39 REKTPSD---LSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYF 95
R+ P D ++ + ++S N++ + L+D E + +A+ T G+ V +QG G F
Sbjct 115 RKVIPKDYKTMTALAKAISKNVLFAHLDDNERSDIFDAMFPVTHIAGETVIQQGNEGDNF 174
Query 96 FIVHSGEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRE 155
++V GE +V VN + V I G +FGE++LI+ + R AT+K + D LWG+ R +R
Sbjct 175 YVVDQGEVDVYVNGEWVTNISEGGSFGELALIYGTPRAATVKAKT-DLKLWGIDRDSYRR 233
Query 156 TL 157
L
Sbjct 234 IL 235
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 46/90 (51%), Gaps = 6/90 (6%)
Query 53 LSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVI----VN 108
LS + SL E +A+A+E F+ G+ + QGE G F+I+ G V+ N
Sbjct 250 LSKVSILESLEKWERLTVADALEPVQFEDGEKIVVQGEPGDDFYIITEGTASVLQRRSPN 309
Query 109 DKV--VNKILTGQAFGEISLIHNSARTATI 136
++ V ++ FGEI+L+ N R AT+
Sbjct 310 EEYVEVGRLGPSDYFGEIALLLNRPRAATV 339
> dre:767685 prkar1b, MGC153624, zgc:153624; protein kinase, cAMP-dependent,
regulatory, type I, beta (EC:2.7.11.1); K04739
cAMP-dependent protein kinase regulator
Length=380
Score = 75.5 bits (184), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 40/122 (32%), Positives = 70/122 (57%), Gaps = 4/122 (3%)
Query 39 REKTPSD---LSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYF 95
R+ P D ++ + ++S N++ + L+D E + +A+ T G+ V +QG+ G F
Sbjct 114 RKVIPKDYKTMTALAKAISKNVLFAHLDDNERSDIFDAMFPVTHIAGETVIQQGDEGDNF 173
Query 96 FIVHSGEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRE 155
+++ GE +V VN + V I G +FGE++LI+ + R AT+K + D LWG+ R +R
Sbjct 174 YVIDQGEVDVYVNGEWVTSIGEGGSFGELALIYGTPRAATVKAKT-DLKLWGIDRDSYRR 232
Query 156 TL 157
L
Sbjct 233 IL 234
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 47/90 (52%), Gaps = 6/90 (6%)
Query 53 LSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIV----N 108
LS + SL+ E +A+A+E F+ G+ + QGE G FFI+ G V+ N
Sbjct 249 LSKVSILESLDKWERLTVADALEPVQFEDGEKIVVQGEPGDDFFIITEGTASVLQRRSDN 308
Query 109 DKV--VNKILTGQAFGEISLIHNSARTATI 136
++ V ++ FGEI+L+ N R AT+
Sbjct 309 EEYVEVGRLGPSDYFGEIALLLNRPRAATV 338
> xla:444805 prkar1b, MGC82149, prkar1; protein kinase, cAMP-dependent,
regulatory, type I,beta; K04739 cAMP-dependent protein
kinase regulator
Length=381
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 38/122 (31%), Positives = 70/122 (57%), Gaps = 4/122 (3%)
Query 39 REKTPSD---LSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYF 95
R+ P D ++ + ++ N++ + L+D E + +A+ T+ G+ V +QG+ G F
Sbjct 115 RKVIPKDYKTMAALAKAIEKNVLFAHLDDNERSDIFDAMFSVTYIAGETVIQQGDEGDNF 174
Query 96 FIVHSGEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRE 155
++V GE +V VN+ + I G +FGE++LI+ + R AT+K + + LWG+ R +R
Sbjct 175 YVVDQGEMDVYVNNDWMTSIGEGGSFGELALIYGTPRAATVKAKT-NVKLWGIDRDSYRR 233
Query 156 TL 157
L
Sbjct 234 IL 235
Score = 45.4 bits (106), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 46/90 (51%), Gaps = 6/90 (6%)
Query 53 LSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIV----N 108
LS + SL+ E +A+A+E F+ G + QGE G FFI+ G V+ N
Sbjct 250 LSKVSILESLDKWERLTVADALEPVQFEDGQKIVVQGEPGDEFFIILEGSAAVLQRRSEN 309
Query 109 DKV--VNKILTGQAFGEISLIHNSARTATI 136
++ V ++ FGEI+L+ N R AT+
Sbjct 310 EEFVEVGRLGPSDYFGEIALLMNRPRAATV 339
> dre:550427 prkar1ab, zgc:112145; protein kinase, cAMP-dependent,
regulatory, type I, alpha (tissue specific extinguisher
1) b (EC:2.7.11.1); K04739 cAMP-dependent protein kinase regulator
Length=379
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 37/122 (30%), Positives = 70/122 (57%), Gaps = 4/122 (3%)
Query 39 REKTPSD---LSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYF 95
R+ P D ++ + ++ N++ S L+D E + +A+ T+ G++V +QG+ G F
Sbjct 113 RKVIPKDYKTMAALAKAIEKNVLFSHLDDNERSDIFDAMFPVTYIAGEIVIQQGDEGDNF 172
Query 96 FIVHSGEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRE 155
+++ GE +V VN + I G +FGE++LI+ + R AT++ + + LWG+ R +R
Sbjct 173 YVIDQGEMDVYVNSEWATSIGEGGSFGELALIYGTPRAATVRAKT-NVKLWGIDRDSYRR 231
Query 156 TL 157
L
Sbjct 232 IL 233
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 45/90 (50%), Gaps = 6/90 (6%)
Query 53 LSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIV----N 108
LS + SL+ E +A+A+E F+ + QGE G FFI+ G V+ N
Sbjct 248 LSKVSILESLDKWERLTVADALEPVQFEDSQKIVVQGEPGDEFFIILEGCAAVLQRRSEN 307
Query 109 DKV--VNKILTGQAFGEISLIHNSARTATI 136
++ V ++ FGEI+L+ N R AT+
Sbjct 308 EEFVEVGRLGPSDYFGEIALLMNRPRAATV 337
> dre:494533 prkar1aa, im:7047729, prkar1a, zgc:92515; protein
kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific
extinguisher 1) a
Length=379
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 37/122 (30%), Positives = 71/122 (58%), Gaps = 4/122 (3%)
Query 39 REKTPSD---LSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYF 95
R+ P D ++ + ++ N++ + L+D E + +A+ T+ G+ V +QG+ G F
Sbjct 113 RKVIPKDYKTMAALAKAIEKNVLFAHLDDNERSDIFDAMFSVTYIAGETVIQQGDEGDNF 172
Query 96 FIVHSGEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRE 155
+++ GE +V VN++ V I G +FGE++LI+ + R AT++ + + LWG+ R +R
Sbjct 173 YVIDQGEMDVYVNNEWVTSIGEGGSFGELALIYGTPRAATVRAKT-NVKLWGIDRDSYRR 231
Query 156 TL 157
L
Sbjct 232 IL 233
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 46/90 (51%), Gaps = 6/90 (6%)
Query 53 LSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIV----N 108
LS + SL+ E +A+A+E F+ G + QG+ G FFI+ G V+ N
Sbjct 248 LSKVSILESLDKWERLTVADALETVQFEDGQKIVVQGQPGDEFFIILEGSAAVLQRRSEN 307
Query 109 DKV--VNKILTGQAFGEISLIHNSARTATI 136
++ V ++ FGEI+L+ N R AT+
Sbjct 308 EEFVEVGRLAPSDYFGEIALLMNRPRAATV 337
> dre:100329979 protein kinase, cAMP-dependent, regulatory, type
I, alpha (tissue specific extinguisher 1) a-like; K04739
cAMP-dependent protein kinase regulator
Length=379
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 37/122 (30%), Positives = 71/122 (58%), Gaps = 4/122 (3%)
Query 39 REKTPSD---LSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYF 95
R+ P D ++ + ++ N++ + L+D E + +A+ T+ G+ V +QG+ G F
Sbjct 113 RKVIPKDYKTMAALAKAIEKNVLFAHLDDNERSDIFDAMFSVTYIAGETVIQQGDEGDNF 172
Query 96 FIVHSGEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRE 155
+++ GE +V VN++ V I G +FGE++LI+ + R AT++ + + LWG+ R +R
Sbjct 173 YVIDQGEMDVYVNNEWVTSIGEGGSFGELALIYGTPRAATVRAKT-NVKLWGIDRDSYRR 231
Query 156 TL 157
L
Sbjct 232 IL 233
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 46/90 (51%), Gaps = 6/90 (6%)
Query 53 LSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIV----N 108
LS + SL+ E +A+A+E F+ G + QG+ G FFI+ G V+ N
Sbjct 248 LSKVSILESLDKWERLTVADALETVQFEDGQKIVVQGQPGDEFFIILEGSAAVLQRRSEN 307
Query 109 DKV--VNKILTGQAFGEISLIHNSARTATI 136
++ V ++ FGEI+L+ N R AT+
Sbjct 308 EEFVEVGRLAPSDYFGEIALLMNRPRAATV 337
> mmu:19084 Prkar1a, 1300018C22Rik, RIalpha, Tse-1, Tse1; protein
kinase, cAMP dependent regulatory, type I, alpha (EC:2.7.11.1);
K04739 cAMP-dependent protein kinase regulator
Length=381
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 37/122 (30%), Positives = 70/122 (57%), Gaps = 4/122 (3%)
Query 39 REKTPSD---LSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYF 95
R+ P D ++ + ++ N++ S L+D E + +A+ +F G+ V +QG+ G F
Sbjct 115 RKVIPKDYKTMAALAKAIEKNVLFSHLDDNERSDIFDAMFPVSFIAGETVIQQGDEGDNF 174
Query 96 FIVHSGEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRE 155
+++ GE +V VN++ + G +FGE++LI+ + R AT+K + + LWG+ R +R
Sbjct 175 YVIDQGEMDVYVNNEWATSVGEGGSFGELALIYGTPRAATVKAKT-NVKLWGIDRDSYRR 233
Query 156 TL 157
L
Sbjct 234 IL 235
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 46/90 (51%), Gaps = 6/90 (6%)
Query 53 LSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIV----N 108
LS + SL+ E +A+A+E F+ G + QGE G FFI+ G V+ N
Sbjct 250 LSKVSILESLDKWERLTVADALEPVQFEDGQKIVVQGEPGDEFFIILEGTAAVLQRRSEN 309
Query 109 DKV--VNKILTGQAFGEISLIHNSARTATI 136
++ V ++ FGEI+L+ N R AT+
Sbjct 310 EEFVEVGRLGPSDYFGEIALLMNRPRAATV 339
> mmu:19092 Prkg2, AW212535, CGKII, MGC130517, Prkgr2, cGK-II;
protein kinase, cGMP-dependent, type II (EC:2.7.11.12); K07376
protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=762
Score = 73.9 bits (180), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 36/131 (27%), Positives = 73/131 (55%), Gaps = 7/131 (5%)
Query 41 KTPSDLSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHS 100
K S+ LI D+L+ N L+ +++ + + +++G V KQGE G++ F++
Sbjct 151 KDSSEKKLITDALNKNQFLKRLDPQQIKDMVECMYGRNYQQGSYVIKQGEPGNHIFVLAE 210
Query 101 GEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETLKQL 160
G EV +K+++ I FGE+++++N RTA++K ++ + W + R+VF+ +++
Sbjct 211 GRLEVFQGEKLLSSIPMWTTFGELAILYNCTRTASVKAIT-NVKTWALDREVFQNIMRRT 269
Query 161 SS------RNF 165
+ RNF
Sbjct 270 AQARDEEYRNF 280
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 30/114 (26%), Positives = 60/114 (52%), Gaps = 6/114 (5%)
Query 50 RDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVN- 108
R+ L + + +L + ++ + + +E + KGD + ++GE GS FFI+ G+ +V +
Sbjct 278 RNFLRSVSLLKNLPEDKLTKIIDCLEVEYYDKGDYIIREGEEGSTFFILAKGKVKVTQST 337
Query 109 -----DKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETL 157
+++ + G+ FGE +LI + R+A I D A + R+ F +T+
Sbjct 338 EGHDQPQLIKTLQKGEYFGEKALISDDVRSANIIAEENDVACLVIDRETFNQTV 391
> hsa:5573 PRKAR1A, CAR, CNC, CNC1, DKFZp779L0468, MGC17251, PKR1,
PPNAD1, PRKAR1, TSE1; protein kinase, cAMP-dependent, regulatory,
type I, alpha (tissue specific extinguisher 1) (EC:2.7.11.1);
K04739 cAMP-dependent protein kinase regulator
Length=381
Score = 73.2 bits (178), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 37/122 (30%), Positives = 70/122 (57%), Gaps = 4/122 (3%)
Query 39 REKTPSD---LSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYF 95
R+ P D ++ + ++ N++ S L+D E + +A+ +F G+ V +QG+ G F
Sbjct 115 RKVIPKDYKTMAALAKAIEKNVLFSHLDDNERSDIFDAMFSVSFIAGETVIQQGDEGDNF 174
Query 96 FIVHSGEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRE 155
+++ GE +V VN++ + G +FGE++LI+ + R AT+K + + LWG+ R +R
Sbjct 175 YVIDQGETDVYVNNEWATSVGEGGSFGELALIYGTPRAATVKAKT-NVKLWGIDRDSYRR 233
Query 156 TL 157
L
Sbjct 234 IL 235
Score = 45.4 bits (106), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 46/90 (51%), Gaps = 6/90 (6%)
Query 53 LSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIV----N 108
LS + SL+ E +A+A+E F+ G + QGE G FFI+ G V+ N
Sbjct 250 LSKVSILESLDKWERLTVADALEPVQFEDGQKIVVQGEPGDEFFIILEGSAAVLQRRSEN 309
Query 109 DKV--VNKILTGQAFGEISLIHNSARTATI 136
++ V ++ FGEI+L+ N R AT+
Sbjct 310 EEFVEVGRLGPSDYFGEIALLMNRPRAATV 339
> hsa:5593 PRKG2, PRKGR2, cGKII; protein kinase, cGMP-dependent,
type II (EC:2.7.11.12); K07376 protein kinase, cGMP-dependent
[EC:2.7.11.12]
Length=762
Score = 70.1 bits (170), Expect = 3e-12, Method: Composition-based stats.
Identities = 35/131 (26%), Positives = 73/131 (55%), Gaps = 7/131 (5%)
Query 41 KTPSDLSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHS 100
K S+ LI D+L+ N L+ +++ + + +++G + KQGE G++ F++
Sbjct 151 KDSSEKKLITDALNKNQFLKRLDPQQIKDMVECMYGRNYQQGSYIIKQGEPGNHIFVLAE 210
Query 101 GEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETLKQL 160
G EV +K+++ I FGE+++++N RTA++K ++ + W + R+VF+ +++
Sbjct 211 GRLEVFQGEKLLSSIPMWTTFGELAILYNCTRTASVKAIT-NVKTWALDREVFQNIMRRT 269
Query 161 SS------RNF 165
+ RNF
Sbjct 270 AQARDEQYRNF 280
Score = 52.8 bits (125), Expect = 5e-07, Method: Composition-based stats.
Identities = 35/139 (25%), Positives = 69/139 (49%), Gaps = 7/139 (5%)
Query 25 TQAEDARLLGHLEKREKTPSDLSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDV 84
T A D + ++ +R D R+ L + + +L + ++ + + +E + KGD
Sbjct 254 TWALDREVFQNIMRRTAQARD-EQYRNFLRSVSLLKNLPEDKLTKIIDCLEVEYYDKGDY 312
Query 85 VTKQGESGSYFFIVHSGEFEVIVND------KVVNKILTGQAFGEISLIHNSARTATIKT 138
+ ++GE GS FFI+ G+ +V + +++ + G+ FGE +LI + R+A I
Sbjct 313 IIREGEEGSTFFILAKGKVKVTQSTEGHDQPQLIKTLQKGEYFGEKALISDDVRSANIIA 372
Query 139 LSEDAALWGVQRQVFRETL 157
D A + R+ F +T+
Sbjct 373 EENDVACLVIDRETFNQTV 391
> dre:100125912 prkg2; protein kinase, cGMP-dependent, type II
(EC:2.7.11.1); K07376 protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=768
Score = 70.1 bits (170), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 35/124 (28%), Positives = 69/124 (55%), Gaps = 7/124 (5%)
Query 48 LIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIV 107
L+ D+L+ N L +V+ + + T+++G+ V KQGE G++ F++ G+ +V
Sbjct 165 LLTDALNKNQYLRRLEVQQVKDMVECMYERTYQQGEYVIKQGEPGNHLFVLADGKLDVYQ 224
Query 108 NDKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETLKQLSS----- 162
++K++ I FGE+++++N RTA++K S + W + R+VF ++ +
Sbjct 225 HNKLLTSIAVWTTFGELAILYNCTRTASVKAAS-NVKTWALDREVFHNIMRMTAEARHEQ 283
Query 163 -RNF 165
RNF
Sbjct 284 YRNF 287
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 28/114 (24%), Positives = 61/114 (53%), Gaps = 7/114 (6%)
Query 50 RDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVN- 108
R+ L + + +SL ++ + + +E + + KGD + ++GE G+ F+I+ +G+ +V +
Sbjct 285 RNFLRSVSLLASLPGDKLTKIVDCLEEY-YNKGDYIIREGEEGNTFYIIANGKIKVTQST 343
Query 109 -----DKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETL 157
+++N + G FGE +LI + R+A I + + R+ F +T+
Sbjct 344 QDHEEPQIINTLGKGDYFGEKALISDDVRSANIIAEEDGVECLVIDRETFSQTV 397
> tgo:TGME49_042070 cAMP-dependent protein kinase regulatory subunit,
putative (EC:2.7.10.2); K04739 cAMP-dependent protein
kinase regulator
Length=308
Score = 69.3 bits (168), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 33/117 (28%), Positives = 69/117 (58%), Gaps = 7/117 (5%)
Query 53 LSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVN---- 108
+ ++ + SSL+ ++E + NA + + KKG V+ +QG+ G +++ +GE +V+
Sbjct 51 IESSFLFSSLDIEDLETVINAFQEVSVKKGTVIIRQGDDGDRLYLIETGEVDVMKKFPGE 110
Query 109 --DKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETLKQLSSR 163
+K + K+ G AFGE++L++N+ R AT+ ++D LW + R F ++ +++
Sbjct 111 KENKFLCKMHPGDAFGELALMYNAPRAATV-IAADDMLLWALDRDSFTNIVRDAAAK 166
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 31/89 (34%), Positives = 52/89 (58%), Gaps = 0/89 (0%)
Query 48 LIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIV 107
+ +SL + ++ E L++A+ T++ GDV+ K+GE+G F+I+ G E I
Sbjct 170 IFEESLKEVRILEDMDPYERSKLSDALRTATYEDGDVIIKEGETGDTFYILLEGAAEAIK 229
Query 108 NDKVVNKILTGQAFGEISLIHNSARTATI 136
NDKVV + G FGE++L+ + R AT+
Sbjct 230 NDKVVMEYKKGGFFGELALLKDQPRAATV 258
> xla:444025 prkg2, MGC82580; protein kinase, cGMP-dependent,
type II (EC:2.7.11.1); K07376 protein kinase, cGMP-dependent
[EC:2.7.11.12]
Length=783
Score = 68.6 bits (166), Expect = 9e-12, Method: Composition-based stats.
Identities = 32/127 (25%), Positives = 71/127 (55%), Gaps = 1/127 (0%)
Query 41 KTPSDLSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHS 100
K S+ LI D+L+ N +++ +++ + + + +G+ + +QGE GS F++
Sbjct 173 KNTSEKILISDALNKNQFTKRMDNHQIQDMVECMYERIYLQGEFIIRQGEPGSQIFVLAD 232
Query 101 GEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETLKQL 160
G+ EV + K++ I FGE+++++N RTA++K ++ + W + R+VF+ +++
Sbjct 233 GKAEVFQDSKLLTSIPVWTTFGELAILYNCTRTASVKAIT-NVRTWALDREVFQNIMRKT 291
Query 161 SSRNFAE 167
+ E
Sbjct 292 AQSKHEE 298
Score = 48.5 bits (114), Expect = 1e-05, Method: Composition-based stats.
Identities = 28/102 (27%), Positives = 52/102 (50%), Gaps = 6/102 (5%)
Query 62 LNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVN------DKVVNKI 115
L D ++ +A+ +E ++ GD + ++GE GS FFI+ G+ +V + + + +
Sbjct 312 LPDHKLMKIADCLELEYYETGDYIIREGEEGSTFFIIAKGKVKVTQSAEGYEEPQYIKLL 371
Query 116 LTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETL 157
G FGE +LI + R+A I E + R+ F +T+
Sbjct 372 EKGDYFGEKALISDDVRSANILAYGERVECLVMDRETFNKTV 413
> cel:C09G4.2 hypothetical protein
Length=617
Score = 68.6 bits (166), Expect = 9e-12, Method: Composition-based stats.
Identities = 30/119 (25%), Positives = 68/119 (57%), Gaps = 1/119 (0%)
Query 41 KTPSDLSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHS 100
K+ + I ++L N +L+ ++E +++A+ G ++ +QG+ GS +++
Sbjct 55 KSDAAFETIGNALRLNSFLRNLDATQIEKISSAMYPVEVPAGAIIIRQGDLGSIMYVIQE 114
Query 101 GEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETLKQ 159
G+ +V+ +++ V + G FGE++++H+ RTAT++ + E LW ++R VF + +
Sbjct 115 GKVQVVKDNRFVRTMEDGALFGELAILHHCERTATVRAI-ESCHLWAIERNVFHAIMME 172
> dre:326020 fd58b04, wu:fd58b04; si:dkey-121j17.5
Length=663
Score = 65.1 bits (157), Expect = 1e-10, Method: Composition-based stats.
Identities = 33/122 (27%), Positives = 66/122 (54%), Gaps = 1/122 (0%)
Query 40 EKTPSDLSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVH 99
+KT S+ S I ++ N L++ ++ + + ++ + GD + K+G G +IV
Sbjct 89 QKTSSETSQIVKAIGKNDFLRRLDEEQISMMVDLMKVLDCRAGDEIIKEGTEGDSMYIVA 148
Query 100 SGEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETLKQ 159
+GE +V + + + G FGE+++++N RTA++K +S LW ++RQ +R +
Sbjct 149 AGELKVTQAGRDLRILSPGDMFGELAILYNCKRTASVKAISA-VKLWCIERQTYRSIMTN 207
Query 160 LS 161
S
Sbjct 208 KS 209
Score = 49.7 bits (117), Expect = 4e-06, Method: Composition-based stats.
Identities = 29/87 (33%), Positives = 49/87 (56%), Gaps = 4/87 (4%)
Query 53 LSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVI--VN-- 108
L T +LND ++ + +++E F+ +V+ ++G G+ F+I+ GE V VN
Sbjct 220 LKTARTLKALNDVQLSKIIDSMEEVRFQDNEVIVREGAEGNTFYIILKGEVLVTKKVNGQ 279
Query 109 DKVVNKILTGQAFGEISLIHNSARTAT 135
K + K+ G+ FGE++LI RTAT
Sbjct 280 QKTIRKMGEGEHFGELALIREILRTAT 306
> cel:F55A8.2 egl-4; EGg Laying defective family member (egl-4);
K07376 protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=780
Score = 65.1 bits (157), Expect = 1e-10, Method: Composition-based stats.
Identities = 41/150 (27%), Positives = 73/150 (48%), Gaps = 3/150 (2%)
Query 11 GERKVQKAIKQQEDTQAEDARLLGHLEKREKTPSDLSLIRDSLSTNLVCSSLNDAEVEAL 70
G ++ +K E T E+ L+ KT +IRD++ N L ++ L
Sbjct 156 GVQRAKKIAVSAEPTNFENKP--ATLQHYNKTVGAKQMIRDAVQKNDFLKQLAKEQIIEL 213
Query 71 ANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKVVNKILTGQAFGEISLIHNS 130
N + + G V ++GE G F+V GE +V ++ K+ G GE+++++N
Sbjct 214 VNCMYEMRARAGQWVIQEGEPGDRLFVVAEGELQVSREGALLGKMRAGTVMGELAILYNC 273
Query 131 ARTATIKTLSEDAALWGVQRQVFRETLKQL 160
RTA+++ L+ D LW + R VF+ ++L
Sbjct 274 TRTASVQALT-DVQLWVLDRSVFQMITQRL 302
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 25/111 (22%), Positives = 53/111 (47%), Gaps = 6/111 (5%)
Query 58 VCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVI------VNDKV 111
+ +L++ + +A+ ++ + G + +QGE G FF+++SG+ +V +
Sbjct 319 IFQNLSEDRISKMADVMDQDYYDGGHYIIRQGEKGDAFFVINSGQVKVTQQIEGETEPRE 378
Query 112 VNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETLKQLSS 162
+ + G FGE +L+ RTA I + + + R+ F + + L S
Sbjct 379 IRVLNQGDFFGERALLGEEVRTANIIAQAPGVEVLTLDRESFGKLIGDLES 429
> cel:R07E4.6 kin-2; protein KINase family member (kin-2); K04739
cAMP-dependent protein kinase regulator
Length=376
Score = 63.5 bits (153), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 32/106 (30%), Positives = 57/106 (53%), Gaps = 1/106 (0%)
Query 52 SLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKV 111
++ NL+ + L + E + + +A+ G+ + +QGE G F+++ G +V VN +
Sbjct 126 AMRKNLLFAHLEEDEQKTMYDAMFPVEKSAGETIIEQGEEGDNFYVIDKGTVDVYVNHEY 185
Query 112 VNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETL 157
V I G +FGE++LI+ + R AT+ D LW + R +R L
Sbjct 186 VLTINEGGSFGELALIYGTPRAATV-IAKTDVKLWAIDRLTYRRIL 230
Score = 45.4 bits (106), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 33/111 (29%), Positives = 54/111 (48%), Gaps = 12/111 (10%)
Query 32 LLGHLEKREKTPSDLSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGES 91
L+G + K+ K + + LS + + L+ E +A+A+E F+ G V +QG+
Sbjct 230 LMGSVTKKRK------MYDEFLSKVQILADLDQWERANVADALERCDFEPGTHVVEQGQP 283
Query 92 GSYFFIVHSGEFEVIVND------KVVNKILTGQAFGEISLIHNSARTATI 136
G FFI+ GE V+ VV + FGEI+L+ + R AT+
Sbjct 284 GDEFFIILEGEANVLQKRSDDAPFDVVGHLGMSDYFGEIALLLDRPRAATV 334
> mmu:19091 Prkg1, AW125416, CGKI, MGC132849, Prkg1b, Prkgr1b;
protein kinase, cGMP-dependent, type I (EC:2.7.11.12); K07376
protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=671
Score = 63.2 bits (152), Expect = 4e-10, Method: Composition-based stats.
Identities = 39/158 (24%), Positives = 76/158 (48%), Gaps = 2/158 (1%)
Query 1 STAAAPHKHGGERKVQ-KAIKQQEDTQAEDARLLGHLEKREKTPSDLSLIRDSLSTNLVC 59
S P H G R + + I + T L K K+ LI++++ N
Sbjct 45 SVLPVPSTHIGPRTTRAQGISAEPQTYRSFHDLRQAFRKFTKSERSKDLIKEAILDNDFM 104
Query 60 SSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKVVNKILTGQ 119
+L ++++ + + + + K + K+G+ GS +++ G+ EV + + G+
Sbjct 105 KNLELSQIQEIVDCMYPVEYGKDSCIIKEGDVGSLVYVMEDGKVEVTKEGVKLCTMGPGK 164
Query 120 AFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETL 157
FGE+++++N RTAT+KTL + LW + RQ F+ +
Sbjct 165 VFGELAILYNCTRTATVKTLV-NVKLWAIDRQCFQTIM 201
Score = 38.9 bits (89), Expect = 0.009, Method: Composition-based stats.
Identities = 24/82 (29%), Positives = 38/82 (46%), Gaps = 6/82 (7%)
Query 61 SLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDK------VVNK 114
SL D + LA+ +E ++ G+ + +QG G FFI+ G+ V D +
Sbjct 224 SLPDEILSKLADVLEETHYENGEYIIRQGARGDTFFIISKGQVNVTREDSPSEDPVFLRT 283
Query 115 ILTGQAFGEISLIHNSARTATI 136
+ G FGE +L RTA +
Sbjct 284 LGKGDWFGEKALQGEDVRTANV 305
> hsa:5592 PRKG1, CGKI, DKFZp686K042, FLJ36117, MGC71944, PGK,
PKG, PRKG1B, PRKGR1B, cGKI-BETA, cGKI-alpha; protein kinase,
cGMP-dependent, type I (EC:2.7.11.12); K07376 protein kinase,
cGMP-dependent [EC:2.7.11.12]
Length=671
Score = 63.2 bits (152), Expect = 4e-10, Method: Composition-based stats.
Identities = 39/158 (24%), Positives = 76/158 (48%), Gaps = 2/158 (1%)
Query 1 STAAAPHKHGGERKVQ-KAIKQQEDTQAEDARLLGHLEKREKTPSDLSLIRDSLSTNLVC 59
S P H G R + + I + T L K K+ LI++++ N
Sbjct 45 SVLPVPSTHIGPRTTRAQGISAEPQTYRSFHDLRQAFRKFTKSERSKDLIKEAILDNDFM 104
Query 60 SSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKVVNKILTGQ 119
+L ++++ + + + + K + K+G+ GS +++ G+ EV + + G+
Sbjct 105 KNLELSQIQEIVDCMYPVEYGKDSCIIKEGDVGSLVYVMEDGKVEVTKEGVKLCTMGPGK 164
Query 120 AFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETL 157
FGE+++++N RTAT+KTL + LW + RQ F+ +
Sbjct 165 VFGELAILYNCTRTATVKTLV-NVKLWAIDRQCFQTIM 201
Score = 35.8 bits (81), Expect = 0.064, Method: Composition-based stats.
Identities = 23/82 (28%), Positives = 37/82 (45%), Gaps = 6/82 (7%)
Query 61 SLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDK------VVNK 114
SL + + LA+ +E ++ G+ + +QG G FFI+ G V D +
Sbjct 224 SLPEEILSKLADVLEETHYENGEYIIRQGARGDTFFIISKGTVNVTREDSPSEDPVFLRT 283
Query 115 ILTGQAFGEISLIHNSARTATI 136
+ G FGE +L RTA +
Sbjct 284 LGKGDWFGEKALQGEDVRTANV 305
> ath:AT2G20050 ATP binding / cAMP-dependent protein kinase regulator/
catalytic/ protein kinase/ protein serine/threonine
phosphatase; K04345 protein kinase A [EC:2.7.11.11]
Length=1094
Score = 62.8 bits (151), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 34/106 (32%), Positives = 58/106 (54%), Gaps = 11/106 (10%)
Query 62 LNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVN----DKVVNKIL- 116
L D++ + L + ++ GD+V KQG G F++V SGEFEV+ + V +IL
Sbjct 495 LTDSQCQVLLDCMQRLEANPGDIVVKQGGEGDCFYVVGSGEFEVLATQDGKNGEVPRILQ 554
Query 117 -----TGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETL 157
+FGE++L+HN A+++ + + LW ++R+ FR L
Sbjct 555 RYTAEKQSSFGELALMHNKPLQASVRAV-DHGTLWALKREDFRGIL 599
> dre:556339 protein kinase, cGMP-dependent, type II-like; K07376
protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=730
Score = 62.8 bits (151), Expect = 6e-10, Method: Composition-based stats.
Identities = 38/155 (24%), Positives = 77/155 (49%), Gaps = 12/155 (7%)
Query 14 KVQKAIKQQEDTQAE-DARLLGHLEKREKTPSDLS----------LIRDSLSTNLVCSSL 62
+V + +K +E AE R H + ++ ++ LI ++L N L
Sbjct 81 EVHRRLKAKEGVSAEPTTRHFCHDHRAHRSSTERQHIRKDSGTKKLINEALMKNDFLKKL 140
Query 63 NDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKVVNKILTGQAFG 122
+ + + + +V ++GE G++ +++ G EVI N K + ++ G AFG
Sbjct 141 EPQHTREMVDCMYEKIYGAEQLVIQEGEPGNFLYVLAEGLLEVIQNGKFLGQMRPGTAFG 200
Query 123 EISLIHNSARTATIKTLSEDAALWGVQRQVFRETL 157
E+++++N RTAT+K +S+ + +W + RQ F+ +
Sbjct 201 ELAILYNCKRTATVKAVSQ-SHIWALDRQTFQTIM 234
Score = 46.2 bits (108), Expect = 5e-05, Method: Composition-based stats.
Identities = 33/107 (30%), Positives = 47/107 (43%), Gaps = 9/107 (8%)
Query 60 SSLNDAEVEALA---NAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEV------IVNDK 110
S L D E LA + +E F KG+ + ++GE G+ FFI+ GE V +
Sbjct 253 SLLKDLPEEKLAKIIDCLEIDYFDKGEYIIREGEEGNTFFIIAKGEVSVTQTTEGFTEPQ 312
Query 111 VVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETL 157
+ + G FGE +LI R+A I D V R F E +
Sbjct 313 EIKTLRVGDYFGEKALISEDVRSANIIAKENDTQCLVVDRDNFNEMV 359
> dre:795338 si:dkey-196h17.4
Length=1669
Score = 61.2 bits (147), Expect = 2e-09, Method: Composition-based stats.
Identities = 43/141 (30%), Positives = 71/141 (50%), Gaps = 4/141 (2%)
Query 21 QQEDTQAEDARLLGHLEKREKTPSDLSLIRDSLSTNLVCSSLNDA-EVEALANAVEFFTF 79
Q+ TQ + LL +E L + +LS + ND + LA V +
Sbjct 989 QRTTTQESEINLLSTVEITRAGKLSLPVFEMTLSKTWLEQLENDGCFIRLLAGGVSQCLY 1048
Query 80 KKGDVVTKQGESGSYFFIVHSGEFEVIVND--KVVNKILTGQAFGEISLIHNSARTATIK 137
+ DV+ K+G+ GS + V GE EV+ N+ KV+ K+ GQ FGE SL+ + R ATI+
Sbjct 1049 RASDVIYKKGDCGSEVYFVEKGEVEVLSNNESKVLFKLSAGQYFGERSLLFHEPRAATIR 1108
Query 138 TLSEDAALWGVQRQVFRETLK 158
+ ++ ++ + ++ E LK
Sbjct 1109 A-ATNSEMYVLSQKSLHEILK 1128
Score = 35.0 bits (79), Expect = 0.13, Method: Composition-based stats.
Identities = 22/77 (28%), Positives = 37/77 (48%), Gaps = 4/77 (5%)
Query 69 ALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKVVN--KILTGQAFGEISL 126
AL+ ++ +T+ G + K GE + + G +V + V +L G FGE+ L
Sbjct 159 ALSLKLKTYTYIPGQTLAKAGEINQNLYYIKHGHVQVFSQNPEVEIATLLPGTLFGEVHL 218
Query 127 IHNSARTATIK--TLSE 141
++ R TI+ TL E
Sbjct 219 LYEIPRNVTIRASTLCE 235
> cel:T20G5.5 epac-1; Exchange Protein Activated by Cyclic AMP
family member (epac-1); K04351 Rap guanine nucleotide exchange
factor (GEF) 4
Length=1234
Score = 60.1 bits (144), Expect = 4e-09, Method: Composition-based stats.
Identities = 31/81 (38%), Positives = 48/81 (59%), Gaps = 0/81 (0%)
Query 82 GDVVTKQGESGSYFFIVHSGEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIKTLSE 141
G VV +QGE G Y++IV G EV VN K+V + G FG+++L+++ R ATI T +
Sbjct 596 GSVVFRQGEIGVYWYIVLKGAVEVNVNGKIVCLLREGDDFGKLALVNDLPRAATIVTYED 655
Query 142 DAALWGVQRQVFRETLKQLSS 162
D+ V + F + L Q+ +
Sbjct 656 DSMFLVVDKHHFNQILHQVEA 676
> dre:394005 prkg1a, MGC63955, prkg1, zgc:63955; protein kinase,
cGMP-dependent, type Ia (EC:2.7.11.1); K07376 protein kinase,
cGMP-dependent [EC:2.7.11.12]
Length=667
Score = 59.3 bits (142), Expect = 6e-09, Method: Composition-based stats.
Identities = 34/131 (25%), Positives = 68/131 (51%), Gaps = 3/131 (2%)
Query 39 REKTPSDLS--LIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFF 96
R SD S LI+ ++ N +L ++++ + + + + K + K+G+ GS +
Sbjct 78 RRVAKSDRSKDLIKSAILDNDFMKNLEMSQIQEIVDCMYPVDYDKNSCIIKEGDVGSLVY 137
Query 97 IVHSGEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRET 156
++ G+ EV + + G+ FGE+++++N RTAT++T+S LW + RQ F+
Sbjct 138 VMEDGKVEVTKEGLKLCTMGPGKVFGELAILYNCTRTATVRTVSS-VKLWAIDRQCFQTI 196
Query 157 LKQLSSRNFAE 167
+ + AE
Sbjct 197 MMRTGLIKHAE 207
Score = 34.3 bits (77), Expect = 0.20, Method: Composition-based stats.
Identities = 23/90 (25%), Positives = 41/90 (45%), Gaps = 6/90 (6%)
Query 53 LSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVND--- 109
L + L L + + LA+ +E ++ G+ + +QG G FFI+ G+ + D
Sbjct 212 LKSVLTFRGLPEEILSKLADVLEETHYEDGNYIIRQGARGDTFFIISKGKVTMTREDCPG 271
Query 110 ---KVVNKILTGQAFGEISLIHNSARTATI 136
+ + G +FGE +L RTA +
Sbjct 272 QEPVYLRSMGRGDSFGEKALQGEDIRTANV 301
> tpv:TP02_0494 cAMP-dependent protein kinase regulatory subunit;
K04739 cAMP-dependent protein kinase regulator
Length=285
Score = 58.9 bits (141), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 37/134 (27%), Positives = 66/134 (49%), Gaps = 12/134 (8%)
Query 37 EKREKTPSDLSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFK------KGDVVTKQGE 90
E EKTP + + I + + + + S + + ++ L A +F K GDV+ KQG+
Sbjct 11 ENYEKTPQEQAKIMEMIKSCFLFSGVTSSGLDLLVKAFDFKAAKIKLFSSAGDVLIKQGD 70
Query 91 SGSYFFIVHSGEFEVIVN-----DKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAAL 145
G +++ SG EV ++ + + G FGE++L++NS R AT+ +E L
Sbjct 71 DGDKLYLIESGTVEVTKKSASGAEEFLCTLTDGDYFGELALMYNSPRAATVVAKTE-MHL 129
Query 146 WGVQRQVFRETLKQ 159
W + R F ++
Sbjct 130 WTLDRTTFNHVVRM 143
Score = 45.8 bits (107), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 23/55 (41%), Positives = 30/55 (54%), Gaps = 0/55 (0%)
Query 83 DVVTKQGESGSYFFIVHSGEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIK 137
+ V KQGE GS F+V G+ E V +K+V G FGEI I R +T+K
Sbjct 185 ETVIKQGEPGSSLFMVLEGQAESFVENKLVKSYNPGDYFGEIGFILKKPRASTVK 239
> mmu:19088 Prkar2b, AI451071, AW061005, Pkarb2, RII(beta); protein
kinase, cAMP dependent regulatory, type II beta (EC:2.7.11.1);
K04739 cAMP-dependent protein kinase regulator
Length=416
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 33/121 (27%), Positives = 65/121 (53%), Gaps = 5/121 (4%)
Query 41 KTPSDLSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHS 100
KT + ++++ L+ +L+ ++ + +A+ K+G+ V QG+ G F+++
Sbjct 135 KTDDQRNRLQEACKDILLFKNLDPEQMSQVLDAMFEKLVKEGEHVIDQGDDGDNFYVIDR 194
Query 101 GEFEVIVNDKVVNKILTGQ----AFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRET 156
G F++ V V + + +FGE++L++N+ R ATI T + ALWG+ R FR
Sbjct 195 GTFDIYVKCDGVGRCVGNYDNRGSFGELALMYNTPRAATI-TATSPGALWGLDRVTFRRI 253
Query 157 L 157
+
Sbjct 254 I 254
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 38/82 (46%), Gaps = 17/82 (20%)
Query 78 TFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDK-----------VVNKILTGQAFGEISL 126
+ G+ + QG+ FFIV SGE ++ + K + + GQ FGE++L
Sbjct 294 VYNDGEQIIAQGDLADSFFIVESGEVKITMKRKGKSEVEENGAVEIARCFRGQYFGELAL 353
Query 127 IHNSARTA------TIKTLSED 142
+ N R A T+K L+ D
Sbjct 354 VTNKPRAASAHAIGTVKCLAMD 375
> pfa:PFL1110c PKAr; CAMP-dependent protein kinase regulatory
subunit, putative; K04739 cAMP-dependent protein kinase regulator
Length=441
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 35/120 (29%), Positives = 64/120 (53%), Gaps = 6/120 (5%)
Query 49 IRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIV- 107
IR++L+ + + + LN E E + NA +KG + +G+ G +++ GE E+
Sbjct 180 IREALNESFLFNHLNKKEFEIIVNAFFDKNVEKGVNIINEGDYGDLLYVIDQGEVEIYKT 239
Query 108 ---NDKVVNKILTGQ-AFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETLKQLSSR 163
N K V +L + FGE++L++NS R AT L++ LW + R+ F +K + ++
Sbjct 240 KENNKKEVLTVLKSKDVFGELALLYNSKRAATATALTK-CHLWALDRESFTYIIKDMVAK 298
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 27/90 (30%), Positives = 48/90 (53%), Gaps = 0/90 (0%)
Query 48 LIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIV 107
+ D LS + ++ E +A+ ++ ++ G+++ K+GE G FFI+ G
Sbjct 302 MYEDILSHVNILKDMDPYERCKVADCLKSKSYNDGEIIIKEGEEGDTFFILIDGNAVASK 361
Query 108 NDKVVNKILTGQAFGEISLIHNSARTATIK 137
++KV+ G FGE++L+ N R ATIK
Sbjct 362 DNKVIKTYTKGDYFGELALLKNKPRAATIK 391
> dre:100329437 protein kinase, cAMP-dependent, regulatory, type
II, alpha-like
Length=385
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 30/118 (25%), Positives = 62/118 (52%), Gaps = 5/118 (4%)
Query 40 EKTPSDLSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVH 99
+KT ++D+ L+ +L + ++ + +++ K G+ + QG+ G F+++
Sbjct 143 QKTDEQRQRLQDACKHILLFKTLEEDQLAEVLDSMFEVLVKPGECIINQGDDGDNFYVIE 202
Query 100 SGEFEVIVNDK----VVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVF 153
G +E+++ V + +FGE++L++N+ R ATI+ L E ALW + R F
Sbjct 203 RGVYEIVIQQDGLQHSVGRYDHKGSFGELALMYNTPRAATIRALQE-GALWALDRATF 259
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 25/96 (26%), Positives = 46/96 (47%), Gaps = 11/96 (11%)
Query 61 SLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKV--------- 111
SL +E + + + +F G+ + +QG+S F+IV SGE +++ K
Sbjct 286 SLQLSERMKIVDVLGMRSFSDGERIIQQGDSADCFYIVESGEVRIMIRSKTRACQLLQEE 345
Query 112 --VNKILTGQAFGEISLIHNSARTATIKTLSEDAAL 145
V + GQ FGE++L+ R A++ + + L
Sbjct 346 VEVARCSRGQYFGELALVTKRPRAASVYAVGDTRCL 381
> hsa:5577 PRKAR2B, PRKAR2, RII-BETA; protein kinase, cAMP-dependent,
regulatory, type II, beta (EC:2.7.11.1); K04739 cAMP-dependent
protein kinase regulator
Length=418
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 33/121 (27%), Positives = 64/121 (52%), Gaps = 5/121 (4%)
Query 41 KTPSDLSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHS 100
KT + ++++ L+ +L+ ++ + +A+ K G+ V QG+ G F+++
Sbjct 137 KTDDQRNRLQEACKDILLFKNLDPEQMSQVLDAMFEKLVKDGEHVIDQGDDGDNFYVIDR 196
Query 101 GEFEVIVNDKVVNKILTGQ----AFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRET 156
G F++ V V + + +FGE++L++N+ R ATI T + ALWG+ R FR
Sbjct 197 GTFDIYVKCDGVGRCVGNYDNRGSFGELALMYNTPRAATI-TATSPGALWGLDRVTFRRI 255
Query 157 L 157
+
Sbjct 256 I 256
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 39/82 (47%), Gaps = 17/82 (20%)
Query 78 TFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDK-----------VVNKILTGQAFGEISL 126
+ G+ + QG+S FFIV SGE ++ + K + + GQ FGE++L
Sbjct 296 VYNDGEQIIAQGDSADSFFIVESGEVKITMKRKGKSEVEENGAVEIARCSRGQYFGELAL 355
Query 127 IHNSARTA------TIKTLSED 142
+ N R A T+K L+ D
Sbjct 356 VTNKPRAASAHAIGTVKCLAMD 377
> dre:405894 prkar2ab, MGC85886, prkar2a, zgc:85886; protein kinase,
cAMP-dependent, regulatory, type II, alpha, B; K04739
cAMP-dependent protein kinase regulator
Length=422
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 30/118 (25%), Positives = 62/118 (52%), Gaps = 5/118 (4%)
Query 40 EKTPSDLSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVH 99
+KT ++D+ L+ +L + ++ + +++ K G+ + QG+ G F+++
Sbjct 143 QKTDEQRQRLQDACKHILLFKTLEEDQLAEVLDSMFEVLVKPGECIINQGDDGDNFYVIE 202
Query 100 SGEFEVIVNDK----VVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVF 153
G +E+++ V + +FGE++L++N+ R ATI+ L E ALW + R F
Sbjct 203 RGVYEIVIQQDGLQHSVGRYDHKGSFGELALMYNTPRAATIRALQE-GALWALDRATF 259
Score = 48.9 bits (115), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 32/119 (26%), Positives = 55/119 (46%), Gaps = 15/119 (12%)
Query 61 SLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKV--------- 111
SL +E + + + +F G+ + +QG+S F+IV SGE +++ K
Sbjct 286 SLQLSERMKIVDVLGMRSFSDGERIIQQGDSADCFYIVESGEVRIMIRSKTRACQLLQEE 345
Query 112 --VNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETL---KQLSSRNF 165
V + GQ FGE++L+ R A++ + + L + Q F L KQ+ RN
Sbjct 346 VEVARCSRGQYFGELALVTKRPRAASVYAVGDTRCLV-IDVQAFERLLGSCKQILKRNI 403
> dre:100332846 acyl-CoA synthetase long-chain family member 6-like
Length=1391
Score = 56.2 bits (134), Expect = 6e-08, Method: Composition-based stats.
Identities = 33/121 (27%), Positives = 66/121 (54%), Gaps = 1/121 (0%)
Query 41 KTPSDLSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHS 100
K+ LI +L+ N L+ +++ L ++ K+G + ++GE G+ +IV
Sbjct 103 KSQESQRLIESALAENECMKYLDRDQLQCLVDSAYPTMLKQGVCLFQEGEHGAQAYIVEE 162
Query 101 GEFEVIVNDKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETLKQL 160
G+ EV + ++ I +G F E+ LIHN + T+ L+ ++ LW ++ QVF++ +++
Sbjct 163 GKLEVSRAGQRLHIIESGMMFEELGLIHNHTCSTTVTALA-NSRLWVMEAQVFQKLMQKS 221
Query 161 S 161
S
Sbjct 222 S 222
Score = 31.2 bits (69), Expect = 1.6, Method: Composition-based stats.
Identities = 22/90 (24%), Positives = 43/90 (47%), Gaps = 7/90 (7%)
Query 79 FKKGDVVTKQGESGSYFFIVHSGEFEV-----IVNDKVVNKILT-GQAFGEISLIHNSAR 132
+ GD + + G FI+ G+ +V + D V+ +LT G +FGE + + R
Sbjct 276 YSDGDYIIQDGSPADKLFILSQGQVKVLEKQCVGEDSVMVCVLTRGDSFGESASQGDDVR 335
Query 133 TATIKTLSEDAALWGVQRQVFRETLKQLSS 162
+ ++ + L + R+VFR+ + L +
Sbjct 336 SISVLAAGDVTCLL-IDREVFRKVTEGLDN 364
> dre:556042 prkar2aa, MGC153742, wu:fb55f12, wu:fj29e12, wu:fj33e06,
zgc:153742; protein kinase, cAMP-dependent, regulatory,
type II, alpha A; K04739 cAMP-dependent protein kinase regulator
Length=397
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 45/80 (56%), Gaps = 7/80 (8%)
Query 83 DVVTKQGESGSYFFIVHSGEFEVIVNDKVVNKILTGQ-----AFGEISLIHNSARTATIK 137
D V QG+ G F+++ G ++++V V GQ +FGE++L++N+ R ATI
Sbjct 161 DHVIDQGDDGDNFYVIERGVYDIVVQKDGVG-CCVGQYNNKGSFGELALMYNTPRAATI- 218
Query 138 TLSEDAALWGVQRQVFRETL 157
++ ALWG+ R FR +
Sbjct 219 IAKDEGALWGLDRATFRRLI 238
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 32/120 (26%), Positives = 55/120 (45%), Gaps = 15/120 (12%)
Query 61 SLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKV--------- 111
SL +E + + + TF+ G+ + QG+ F+IV SGE ++++ K
Sbjct 261 SLELSERMKIVDVLGVKTFQDGERIIMQGDKADCFYIVESGEVKIMMKSKTKADRQDNAE 320
Query 112 --VNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETL---KQLSSRNFA 166
+ + GQ FGE++L+ N R A+ + D + Q F L K++ RN A
Sbjct 321 VEIARCSRGQYFGELALVTNKPRAASAYAIG-DVKCLVIDIQAFERLLGPCKEIMKRNIA 379
> hsa:5576 PRKAR2A, MGC3606, PKR2, PRKAR2; protein kinase, cAMP-dependent,
regulatory, type II, alpha (EC:2.7.11.1); K04739
cAMP-dependent protein kinase regulator
Length=404
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 36/142 (25%), Positives = 73/142 (51%), Gaps = 13/142 (9%)
Query 20 KQQEDTQAEDARLLGHLEKREKTPSDLSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTF 79
+++EDT D R++ KT ++++ L+ +L+ ++ + +A+
Sbjct 109 EEEEDT---DPRVI-----HPKTDEQRCRLQEACKDILLFKNLDQEQLSQVLDAMFERIV 160
Query 80 KKGDVVTKQGESGSYFFIVHSGEFEVIVND----KVVNKILTGQAFGEISLIHNSARTAT 135
K + V QG+ G F+++ G ++++V + V + +FGE++L++N+ R AT
Sbjct 161 KADEHVIDQGDDGDNFYVIERGTYDILVTKDNQTRSVGQYDNRGSFGELALMYNTPRAAT 220
Query 136 IKTLSEDAALWGVQRQVFRETL 157
I SE +LWG+ R FR +
Sbjct 221 IVATSE-GSLWGLDRVTFRRII 241
Score = 42.0 bits (97), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 19/69 (27%), Positives = 34/69 (49%), Gaps = 12/69 (17%)
Query 79 FKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKV------------VNKILTGQAFGEISL 126
+K G+ + QGE F+I+ SGE +++ + + + GQ FGE++L
Sbjct 282 YKDGERIITQGEKADSFYIIESGEVSILIRSRTKSNKDGGNQEVEIARCHKGQYFGELAL 341
Query 127 IHNSARTAT 135
+ N R A+
Sbjct 342 VTNKPRAAS 350
> cpv:cgd7_120 cAMP-dependent protein kinase regulatory subunit
; K04739 cAMP-dependent protein kinase regulator
Length=345
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 33/123 (26%), Positives = 61/123 (49%), Gaps = 6/123 (4%)
Query 49 IRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVN 108
IR+ L + + +SL+D E++ + A + KK + QG++G +I+ G E
Sbjct 90 IREKLLESFMFTSLDDDELKTVILACVETSVKKDTEIITQGDNGDKLYIIDQGVVECYKK 149
Query 109 D-----KVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETLKQLSSR 163
K + + G AFGE++L++N R A++ D LW + R+ F +K +S+
Sbjct 150 TSTEPRKHLCDLNPGDAFGELALLYNCPRAASV-VAKTDCLLWALDRETFNHIVKGSASK 208
Query 164 NFA 166
+
Sbjct 209 RIS 211
Score = 45.8 bits (107), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 25/100 (25%), Positives = 50/100 (50%), Gaps = 1/100 (1%)
Query 58 VCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKVVNKILT 117
+ +++ E+ L ++ F+ G + KQGE G F+++ +G + ++ V
Sbjct 222 ILKTMDVYELNKLTMVLKSSIFEDGQEIIKQGEQGDTFYLIITGNAVALKDNVEVMSYKR 281
Query 118 GQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETL 157
G FGE++L+ N+ R AT+K + + R+ F+ L
Sbjct 282 GDYFGELALLRNAPRAATVKARGRCKVAY-LDRKAFKRVL 320
> xla:414594 prkar2b, MGC83177; protein kinase, cAMP-dependent,
regulatory, type II, beta; K04739 cAMP-dependent protein kinase
regulator
Length=402
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 44/80 (55%), Gaps = 5/80 (6%)
Query 82 GDVVTKQGESGSYFFIVHSGEFEVIVNDKVVNKILTGQ----AFGEISLIHNSARTATIK 137
G+ V QG+ G F+++ G +++ V V + + +FGE++L++N+ R ATI
Sbjct 162 GEHVIDQGDDGDNFYVIDRGTYDIFVKSDGVARCVCAYDNRGSFGELALMYNTPRAATIV 221
Query 138 TLSEDAALWGVQRQVFRETL 157
S A+WG+ R FR +
Sbjct 222 ATSV-GAIWGLDRATFRRII 240
Score = 38.5 bits (88), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 33/69 (47%), Gaps = 11/69 (15%)
Query 78 TFKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKV-----------VNKILTGQAFGEISL 126
T+ + + QG+ FFIV SGE + + K + + + GQ FGE++L
Sbjct 280 TYCDEEQIIAQGDGADSFFIVESGEVRITMKSKSKPDAETNEAVEIARYIRGQYFGELAL 339
Query 127 IHNSARTAT 135
+ N R A+
Sbjct 340 VTNKPRAAS 348
> mmu:19087 Prkar2a, 1110061A24Rik, AI317181, AI836829, RII(alpha);
protein kinase, cAMP dependent regulatory, type II alpha
(EC:2.7.11.1); K04739 cAMP-dependent protein kinase regulator
Length=402
Score = 52.8 bits (125), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 44/77 (57%), Gaps = 5/77 (6%)
Query 85 VTKQGESGSYFFIVHSGEFEVIVND----KVVNKILTGQAFGEISLIHNSARTATIKTLS 140
V QG+ G F+++ G ++++V + V + +FGE++L++N+ R ATI S
Sbjct 164 VIDQGDDGDNFYVIERGTYDILVTKDNQTRSVGQYDNRGSFGELALMYNTPRAATIIATS 223
Query 141 EDAALWGVQRQVFRETL 157
E +LWG+ R FR +
Sbjct 224 E-GSLWGLDRVTFRRII 239
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 33/69 (47%), Gaps = 12/69 (17%)
Query 79 FKKGDVVTKQGESGSYFFIVHSGEFEVIVNDKV------------VNKILTGQAFGEISL 126
+K G+ + QGE F+I+ SGE +++ K + GQ FGE++L
Sbjct 280 YKDGERIIAQGEKADSFYIIESGEVSILIRSKTKSNKNGGNQEVEIAHCHKGQYFGELAL 339
Query 127 IHNSARTAT 135
+ N R A+
Sbjct 340 VTNKPRAAS 348
> tgo:TGME49_111300 cAMP-dependent protein kinase regulatory subunit,
putative (EC:2.7.11.12); K04739 cAMP-dependent protein
kinase regulator
Length=410
Score = 52.4 bits (124), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 29/115 (25%), Positives = 64/115 (55%), Gaps = 5/115 (4%)
Query 53 LSTNLVCSSLNDAEVEALANAVEFFTFKKGDVVTKQGESGSYFFIVHSGEFEV--IVN-- 108
L + + +SL++ ++ + A++ + + ++G+ G +IV SGE +++
Sbjct 160 LRQSFLFNSLDEKDLNTVILAMQEKKIEASTCLIREGDDGECLYIVQSGELNCSKLIDGE 219
Query 109 DKVVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETLKQLSSR 163
++VV + G AFGE++L++N+ R AT+ ++S LW + R F +K +++
Sbjct 220 ERVVKVVGPGDAFGELALLYNAPRAATVTSVSA-CDLWELGRDTFNAIVKDAATK 273
Score = 37.0 bits (84), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 33/73 (45%), Gaps = 6/73 (8%)
Query 70 LANAVEFFTFKKGDVVTKQGESGSYFFIVHSG------EFEVIVNDKVVNKILTGQAFGE 123
+A+A+ F G + +QGE G F+IV G F V K G FGE
Sbjct 299 VADALRTEMFTDGAYIVRQGELGDVFYIVEEGSAVATKSFGPGQPPIEVKKYQAGDYFGE 358
Query 124 ISLIHNSARTATI 136
++LI+ R A +
Sbjct 359 LALINEEPRAANV 371
> dre:100329672 cAMP-regulated guanine nucleotide exchange factor
II-like; K04351 Rap guanine nucleotide exchange factor (GEF)
4
Length=1005
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 34/125 (27%), Positives = 64/125 (51%), Gaps = 2/125 (1%)
Query 40 EKTPSDLSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKK-GDVVTKQGESGSYFFIV 98
++T DL +I D L S L++ LA + F + K G V+ QGE G+ ++I+
Sbjct 336 QRTAEDLEIIYDELLHIKAVSHLSNTVKRELAGVLIFESHAKAGTVLFNQGEEGTSWYII 395
Query 99 HSGEFEVIVNDK-VVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETL 157
G V++ K VV + G FG+++L++++ R A+I ++ V ++ F L
Sbjct 396 QKGSVNVVIYGKGVVCTLHEGDDFGKLALVNDAPRAASIVLREDNCHFLRVDKEDFNRIL 455
Query 158 KQLSS 162
+ + +
Sbjct 456 RDVEA 460
Score = 35.8 bits (81), Expect = 0.066, Method: Compositional matrix adjust.
Identities = 25/87 (28%), Positives = 50/87 (57%), Gaps = 10/87 (11%)
Query 76 FFTF-KKGDVVTKQGESGSYFFIVHSGEFEVIVNDK-------VVNKILTGQAFGEISLI 127
F+ + +KG + +QG+ G+ ++ V SG +V V++ + + G AFGE S++
Sbjct 61 FYEYLEKGITLYRQGDIGTSWYAVLSGSLDVKVSETANHQDAVTICTLGVGTAFGE-SIL 119
Query 128 HNSARTATIKTLSEDAALWGVQRQVFR 154
N+ R ATI + E++ L ++++ F+
Sbjct 120 DNTPRHATIVS-RENSELLRIEQREFK 145
> hsa:10411 RAPGEF3, CAMP-GEFI, EPAC, EPAC1, HSU79275, MGC21410,
bcm910; Rap guanine nucleotide exchange factor (GEF) 3; K08014
Rap guanine nucleotide exchange factor (GEF) 3
Length=923
Score = 48.9 bits (115), Expect = 8e-06, Method: Composition-based stats.
Identities = 32/126 (25%), Positives = 65/126 (51%), Gaps = 2/126 (1%)
Query 40 EKTPSDLSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKK-GDVVTKQGESGSYFFIV 98
++T +L LI + L + L+++ LA + F K G V+ QG+ G+ ++I+
Sbjct 227 QRTDEELDLIFEELLHIKAVAHLSNSVKRELAAVLLFEPHSKAGTVLFSQGDKGTSWYII 286
Query 99 HSGEFEVIVNDK-VVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETL 157
G V+ + K +V + G FG+++L++++ R ATI ++ V +Q F +
Sbjct 287 WKGSVNVVTHGKGLVTTLHEGDDFGQLALVNDAPRAATIILREDNCHFLRVDKQDFNRII 346
Query 158 KQLSSR 163
K + ++
Sbjct 347 KDVEAK 352
> mmu:56508 Rapgef4, 1300003D15Rik, 5730402K07Rik, 6330581N18Rik,
EPAC_2, Epac-2, Epac2, KIAA4040, mKIAA4040; Rap guanine
nucleotide exchange factor (GEF) 4; K04351 Rap guanine nucleotide
exchange factor (GEF) 4
Length=1011
Score = 48.9 bits (115), Expect = 9e-06, Method: Composition-based stats.
Identities = 34/125 (27%), Positives = 63/125 (50%), Gaps = 2/125 (1%)
Query 40 EKTPSDLSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKKG-DVVTKQGESGSYFFIV 98
++T DL +I D L S L+ LA + F + KG V+ QGE G+ ++I+
Sbjct 338 QRTVDDLEIIYDELLHIKALSHLSTTVKRELAGVLIFESHAKGGTVLFNQGEEGTSWYII 397
Query 99 HSGEFEVIVNDK-VVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETL 157
G V++ K VV + G FG+++L++++ R A+I ++ V ++ F L
Sbjct 398 LKGSVNVVIYGKGVVCTLHEGDDFGKLALVNDAPRAASIVLREDNCHFLRVDKEDFNRIL 457
Query 158 KQLSS 162
+ + +
Sbjct 458 RDVEA 462
Score = 33.5 bits (75), Expect = 0.33, Method: Composition-based stats.
Identities = 24/83 (28%), Positives = 46/83 (55%), Gaps = 9/83 (10%)
Query 79 FKKGDVVTKQGESGSYFFIVHSGEFEVIVNDK-------VVNKILTGQAFGEISLIHNSA 131
+KG + +QG+ G+ ++ V +G +V V++ + + G AFGE S++ N+
Sbjct 64 LEKGITLFRQGDIGTNWYAVLAGSLDVKVSETSSHQDAVTICTLGIGTAFGE-SILDNTP 122
Query 132 RTATIKTLSEDAALWGVQRQVFR 154
R ATI T E + L ++++ F+
Sbjct 123 RHATIVT-RESSELLRIEQEDFK 144
> mmu:223864 Rapgef3, 2310016P22Rik, 9330170P05Rik, Epac, Epac1,
MGC19192; Rap guanine nucleotide exchange factor (GEF) 3;
K08014 Rap guanine nucleotide exchange factor (GEF) 3
Length=926
Score = 47.8 bits (112), Expect = 2e-05, Method: Composition-based stats.
Identities = 32/126 (25%), Positives = 64/126 (50%), Gaps = 2/126 (1%)
Query 40 EKTPSDLSLIRDSLSTNLVCSSLNDAEVEALANAVEFFTFKK-GDVVTKQGESGSYFFIV 98
++T +L LI + L + L+++ LA + F K G V+ QG+ G+ ++I+
Sbjct 227 QRTDEELDLIFEELLHIKAVAHLSNSVKRELAAVLLFEPHSKAGTVLFSQGDKGTSWYII 286
Query 99 HSGEFEVIVNDK-VVNKILTGQAFGEISLIHNSARTATIKTLSEDAALWGVQRQVFRETL 157
G V+ + K +V + G FG+++L++++ R ATI + V +Q F +
Sbjct 287 WKGSVNVVTHGKGLVTTLHEGDDFGQLALVNDAPRAATIILRENNCHFLRVDKQDFNRII 346
Query 158 KQLSSR 163
K + ++
Sbjct 347 KDVEAK 352
> dre:100331387 cAMP-dependent protein kinase, regulatory subunit
beta 2-like; K04739 cAMP-dependent protein kinase regulator
Length=403
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 24/80 (30%), Positives = 42/80 (52%), Gaps = 5/80 (6%)
Query 82 GDVVTKQGESGSYFFIVHSGEFEVIV----NDKVVNKILTGQAFGEISLIHNSARTATIK 137
G+ + Q + G F+++ G FE+++ + V +FGE++L++N+ R ATI
Sbjct 163 GEHIIDQDDDGDNFYVIERGMFEILLKADGTTRTVGSYDNRGSFGELALMYNTPRAATII 222
Query 138 TLSEDAALWGVQRQVFRETL 157
S ALW + R FR +
Sbjct 223 ATSP-GALWCLDRLTFRRII 241
Lambda K H
0.313 0.128 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4157683456
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40