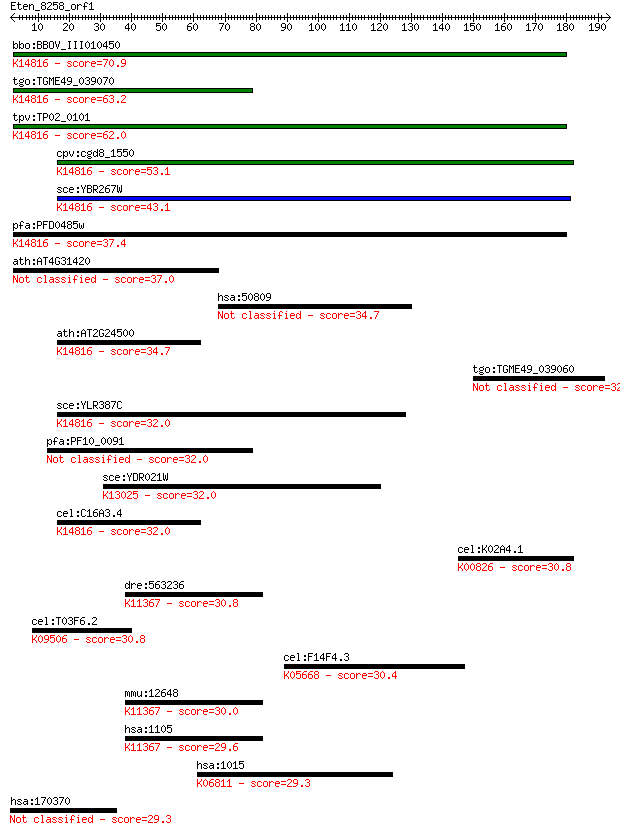
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8258_orf1
Length=193
Score E
Sequences producing significant alignments: (Bits) Value
bbo:BBOV_III010450 17.m07902; hypothetical protein; K14816 pre... 70.9 2e-12
tgo:TGME49_039070 hypothetical protein ; K14816 pre-60S factor... 63.2 6e-10
tpv:TP02_0101 hypothetical protein; K14816 pre-60S factor REI1 62.0 1e-09
cpv:cgd8_1550 S. cerevisiae YLR387c-like protein with 2x C2H2 ... 53.1 6e-07
sce:YBR267W REI1; Cytoplasmic pre-60S factor; required for the... 43.1 6e-04
pfa:PFD0485w conserved Plasmodium protein, unknown function; K... 37.4 0.033
ath:AT4G31420 zinc finger (C2H2 type) family protein 37.0 0.041
hsa:50809 HP1BP3, HP1-BP74, MGC43701; heterochromatin protein ... 34.7 0.19
ath:AT2G24500 FZF; FZF; transcription factor; K14816 pre-60S f... 34.7 0.21
tgo:TGME49_039060 hypothetical protein 32.7 0.90
sce:YLR387C REH1; Cytoplasmic 60S subunit biogenesis factor, a... 32.0 1.2
pfa:PF10_0091 zinc finger protein, putative 32.0 1.3
sce:YDR021W FAL1; Fal1p (EC:3.6.1.-); K13025 ATP-dependent RNA... 32.0 1.4
cel:C16A3.4 hypothetical protein; K14816 pre-60S factor REI1 32.0 1.5
cel:K02A4.1 bcat-1; Branched Chain AminoTransferase family mem... 30.8 3.1
dre:563236 MGC173506, wu:fc26h11, wu:fk85d05; zgc:173506; K113... 30.8 3.1
cel:T03F6.2 dnj-17; DNaJ domain (prokaryotic heat shock protei... 30.8 3.4
cel:F14F4.3 mrp-5; Multidrug Resistance Protein family member ... 30.4 4.2
mmu:12648 Chd1, 4930525N21Rik, AI851787, AW555109, MGC141554; ... 30.0 5.2
hsa:1105 CHD1, DKFZp686E2337; chromodomain helicase DNA bindin... 29.6 6.2
hsa:1015 CDH17, CDH16, FLJ26931, HPT-1, HPT1, MGC138218, MGC14... 29.3 9.6
hsa:170370 FAM170B, C10orf73; family with sequence similarity ... 29.3 9.8
> bbo:BBOV_III010450 17.m07902; hypothetical protein; K14816 pre-60S
factor REI1
Length=387
Score = 70.9 bits (172), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 47/178 (26%), Positives = 81/178 (45%), Gaps = 32/178 (17%)
Query 2 LLRCLWQEQQKQPRCLFCCRGFKGIRAALQHMQQQRHFQLKWDEEQQDLLHRFYDYKKSY 61
LL L+ + Q C++C + F I A HM+Q++H +L +++ D + +FYD+ +SY
Sbjct 180 LLGYLYDKIHNQYTCIYCHKPFGSIYAVNHHMEQKQHRKL--NDDDLDEVAQFYDFTRSY 237
Query 62 YEILGRLPEVNNAQLVLPDNLSSSIQDATSPLAAAKRTGRPLEAEAEVVAGEDEGDWEDC 121
E++ + D S++ S +DE DWED
Sbjct 238 AELM-----IKGVDYYKHDGTESTVDRDDSEYT------------------DDEDDWEDV 274
Query 122 SSDEEGSANAASEQRRLEEMLQARGWRHARVTDEGNLQLPSGQEVLHRSHAIFCRQRV 179
+ E +EQ + L + G AR++ GNL LP+G+E +HR + +Q +
Sbjct 275 ITPE-------TEQHDAIQTLSSLGLYRARISPSGNLTLPNGREAIHRDISYVYKQHL 325
> tgo:TGME49_039070 hypothetical protein ; K14816 pre-60S factor
REI1
Length=429
Score = 63.2 bits (152), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 45/81 (55%), Gaps = 4/81 (4%)
Query 2 LLRCLWQEQQKQPRCLFCCRGFKGIRAALQHMQQQRHFQLKWDEEQQDLLHRF----YDY 57
LR +W+ Q ++PRCL+C + F + AA QHMQ + H QL+W + L R +D+
Sbjct 262 FLRIIWKAQMRKPRCLWCMQRFASVEAAQQHMQSKGHTQLRWADSADSALQRALEPCFDF 321
Query 58 KKSYYEILGRLPEVNNAQLVL 78
+ SY +L R ++ Q L
Sbjct 322 RASYLALLERRAQLAETQKAL 342
> tpv:TP02_0101 hypothetical protein; K14816 pre-60S factor REI1
Length=365
Score = 62.0 bits (149), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 44/178 (24%), Positives = 82/178 (46%), Gaps = 37/178 (20%)
Query 2 LLRCLWQEQQKQPRCLFCCRGFKGIRAALQHMQQQRHFQLKWDEEQQDLLHRFYDYKKSY 61
LLR L + + +CL+C + F A L HM ++H ++ ++++ D + FYD+ SY
Sbjct 163 LLRYLHNKIYHENKCLYCDKPFLDHYATLHHMVDKQHHKI--NDDRFDEISSFYDFIDSY 220
Query 62 YEILGRLPEVNNAQLVLPDNLSSSIQDATSPLAAAKRTGRPLEAEAEVVAGEDEGDWEDC 121
++ + + K + + + G+DE DWED
Sbjct 221 VNLI---------------------------VDSKKSSSSDGSSSYKTQEGQDE-DWEDI 252
Query 122 SSDEEGSANAASEQRRLEEMLQARGWRHARVTDEGNLQLPSGQEVLHRSHAIFCRQRV 179
S S N+ + +E +L + G++ A + + GNL LP+G+E +HR + +Q +
Sbjct 253 FS----STNSPTS---VENLLSSYGFKKAYIMENGNLSLPNGKEAVHRELSYVYKQNL 303
> cpv:cgd8_1550 S. cerevisiae YLR387c-like protein with 2x C2H2
like zinc fingers conserved across eukaryotes plus an apicomplexan-specific
globular domain ; K14816 pre-60S factor REI1
Length=392
Score = 53.1 bits (126), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 42/167 (25%), Positives = 80/167 (47%), Gaps = 20/167 (11%)
Query 16 CLFCCRGFKGIRAALQHMQQQRHFQLKWD-EEQQDLLHRFYDYKKSYYEILGRLPEVNNA 74
CL+C + F +RA HM H L + Q++ L FY+Y SY E++ P N
Sbjct 165 CLYCDKIFSSLRAVRDHMISLGHTMLGTHLDIQKEELESFYNYSLSYKELI---PNFN-- 219
Query 75 QLVLPDNLSSSIQDATSPLAAAKRTGRPLEAEAEVVAGEDEGDWEDCSSDEEGSANAASE 134
+L + D++ +++ + + + + +G +++E N +
Sbjct 220 KLSITDDV---LKEENNDDWEYIDEEEEEDDDDDDFSG---------NTNEIRKLNK--K 265
Query 135 QRRLEEMLQARGWRHARVTDEGNLQLPSGQEVLHRSHAIFCRQRVRR 181
+ L+E+L +T+ G+L+LP+G+EV+HR+ A +QR+ R
Sbjct 266 EMTLDEILSMYNLSKPEITEFGDLRLPNGKEVVHRNLAYIYKQRIPR 312
> sce:YBR267W REI1; Cytoplasmic pre-60S factor; required for the
correct recycling of shuttling factors Alb1, Arx1 and Tif6
at the end of the ribosomal large subunit biogenesis; involved
in bud growth in the mitotic signaling network; K14816
pre-60S factor REI1
Length=393
Score = 43.1 bits (100), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 39/166 (23%), Positives = 62/166 (37%), Gaps = 56/166 (33%)
Query 16 CLFCCRGFKGIRAALQHMQQQRHFQLKWDEEQQDL-LHRFYDYKKSYYEILGRLPEVNNA 74
C+ C + + A QHM +RH ++ ++ E + L + FYD+ SY
Sbjct 217 CIVCNYQGRTLTAVRQHMLAKRHCKIPYESEDERLEISEFYDFTSSY------------- 263
Query 75 QLVLPDNLSSSIQDATSPLAAAKRTGRPLEAEAEVVAGEDEGDWEDCSSDEEGSANAASE 134
++ T+P ++E DWED SDE GS +
Sbjct 264 ---------ANFNSNTTP--------------------DNEDDWEDVGSDEAGSDDEDLP 294
Query 135 QRRLEEMLQARGWRHARVTDEGNLQLPSGQEVLHRSHAIFCRQRVR 180
Q L D L LP+G +V HRS + +Q ++
Sbjct 295 QEYL-------------YNDGIELHLPTGIKVGHRSLQRYYKQDLK 327
> pfa:PFD0485w conserved Plasmodium protein, unknown function;
K14816 pre-60S factor REI1
Length=575
Score = 37.4 bits (85), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 40/224 (17%), Positives = 91/224 (40%), Gaps = 50/224 (22%)
Query 2 LLRCLWQEQQKQPRCLFCCRGFKGIRAALQHMQQQRHFQLKWDEEQQDLLHRFYDYKKSY 61
+L + ++ ++ C++C + K +++ HM + H +L + + ++YD+ K+Y
Sbjct 292 ILLTIGKKIYEENICIYCFKYAKCVKSLQAHMICKSHTKLHTN--FMVYIQKYYDFSKTY 349
Query 62 YEILGRLPEVNN------------------AQLVLPDNLSSSIQD----ATSPLAAAKRT 99
++L + +NN QL + DN + + + + + K+
Sbjct 350 VDLLNKY--INNKQDKKLLLYMLNHEQNKEKQLQIHDNKNQNHHNDNHLENNNVLTKKKL 407
Query 100 GRPLEAEAE-------VVAGEDEGDW-------------EDCSSDE----EGSANAASEQ 135
+ +E ++ E D +D SSD+ E N +
Sbjct 408 SNDTDNNSEDYPNDSYILKKEKNQDTSLSNSYDNHDNTSDDNSSDDYKIKETDLNKNIDY 467
Query 136 RRLEEMLQARGWRHARVTDEGNLQLPSGQEVLHRSHAIFCRQRV 179
++ ++L+ G+ + + NL LP G E ++R A +Q++
Sbjct 468 NKIYQVLEEFGYIKPELNEYNNLILPDGSEAINRKLAYIFKQKL 511
> ath:AT4G31420 zinc finger (C2H2 type) family protein
Length=404
Score = 37.0 bits (84), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 35/72 (48%), Gaps = 6/72 (8%)
Query 2 LLRCLWQEQQKQPRCLFC---CRGFKGIRAALQHMQQQRHFQLKWDEEQQDLLHR---FY 55
LL L + ++ CL+C CR F + A +HM+ + H +L + + + FY
Sbjct 217 LLTYLGLKVKRDFMCLYCNELCRPFSSLEAVRKHMEAKSHCKLHYGDGDDEEDAELEEFY 276
Query 56 DYKKSYYEILGR 67
DY SY + G+
Sbjct 277 DYSSSYVDEAGK 288
> hsa:50809 HP1BP3, HP1-BP74, MGC43701; heterochromatin protein
1, binding protein 3
Length=553
Score = 34.7 bits (78), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 32/62 (51%), Gaps = 0/62 (0%)
Query 68 LPEVNNAQLVLPDNLSSSIQDATSPLAAAKRTGRPLEAEAEVVAGEDEGDWEDCSSDEEG 127
LP + AQL+ D L ++D+T P+ + R ++++ GE+E D SS+E
Sbjct 16 LPLIVGAQLIHADKLGEKVEDSTMPIRRTVNSTRETPPKSKLAEGEEEKPEPDISSEESV 75
Query 128 SA 129
S
Sbjct 76 ST 77
> ath:AT2G24500 FZF; FZF; transcription factor; K14816 pre-60S
factor REI1
Length=395
Score = 34.7 bits (78), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 27/53 (50%), Gaps = 8/53 (15%)
Query 16 CLFC---CRGFKGIRAALQHMQQQRHFQLKW----DEEQQDLLHRFYDYKKSY 61
CL+C C F + A +HM + H ++ + DEE +L FYDY SY
Sbjct 224 CLYCNELCHPFSSLEAVRKHMDAKGHCKVHYGDGGDEEDAEL-EEFYDYSSSY 275
> tgo:TGME49_039060 hypothetical protein
Length=158
Score = 32.7 bits (73), Expect = 0.90, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 29/43 (67%), Gaps = 1/43 (2%)
Query 150 ARVTDEGNLQLPSGQEVLHRSHAIFCRQRV-RRVERQAAEQCV 191
AR+T+ G+L+LP G+E+++R A +QR+ RRV A Q +
Sbjct 3 ARLTETGDLRLPDGRELVNRHVAYIYKQRLGRRVPGDAEAQVL 45
> sce:YLR387C REH1; Cytoplasmic 60S subunit biogenesis factor,
associates with pre-60S particles; similar to Rei1p and shares
partially redundant function in cytoplasmic 60S subunit
maturation; contains dispersed C2H2 zinc finger domains; K14816
pre-60S factor REI1
Length=432
Score = 32.0 bits (71), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 29/115 (25%), Positives = 48/115 (41%), Gaps = 18/115 (15%)
Query 16 CLFCCRGFKGIRAALQHMQQQRHFQLKWD-EEQQDLLHRFYDYKKSYYEILGRLPEVNNA 74
C F G + IRA HM +RH +L ++ +E++ L FYD+ + I L
Sbjct 242 CNFHGSGLESIRA---HMASKRHCRLPYETKEERQLFAPFYDFTYDDHSISKNLQN---- 294
Query 75 QLVLPDNLSSSIQDATSPLAAAKRTGRPLEAEAEVVAGEDE--GDWEDCSSDEEG 127
+I S + AK E + +V+ E++ ++ S DE G
Sbjct 295 --------DRAITSKLSSVYGAKNDEEDGEVDITLVSSENDINANYTTVSIDESG 341
> pfa:PF10_0091 zinc finger protein, putative
Length=362
Score = 32.0 bits (71), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 19/71 (26%), Positives = 35/71 (49%), Gaps = 6/71 (8%)
Query 13 QPRCLFCCRGFKGIRAALQHMQQQRHFQL-----KWDEEQQDLLHRFYDYKKSYYEILGR 67
+P C +C R F + +QH Q+ +HF+ K D ++H +K + +
Sbjct 13 KPFCYYCDREFDDEKILIQH-QKAKHFKCLHCNRKLDMANGLVVHMMQVHKTNLKSVPNA 71
Query 68 LPEVNNAQLVL 78
LP+ N+ +LV+
Sbjct 72 LPKRNDPELVI 82
> sce:YDR021W FAL1; Fal1p (EC:3.6.1.-); K13025 ATP-dependent RNA
helicase [EC:3.6.4.13]
Length=399
Score = 32.0 bits (71), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 44/95 (46%), Gaps = 7/95 (7%)
Query 31 QHMQQQRHFQLKWDEEQQDLLHRFYDYKKSYYEILGRLPEVNNAQLVLPDNLSSSIQDAT 90
+ M Q R+ Q+ +E +LL +K+ Y+I +LP+ N +V+ ++ I + T
Sbjct 159 KQMLQTRNVQMLVLDEADELLSETLGFKQQIYDIFAKLPK-NCQVVVVSATMNKDILEVT 217
Query 91 -----SPLAA-AKRTGRPLEAEAEVVAGEDEGDWE 119
P+ KR LE + V D+ +W+
Sbjct 218 RKFMNDPVKILVKRDEISLEGIKQYVVNVDKEEWK 252
> cel:C16A3.4 hypothetical protein; K14816 pre-60S factor REI1
Length=375
Score = 32.0 bits (71), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 27/49 (55%), Gaps = 3/49 (6%)
Query 16 CLFCCRG---FKGIRAALQHMQQQRHFQLKWDEEQQDLLHRFYDYKKSY 61
C+FC ++ +++ QHM+ ++H +L+ D E L +YDY Y
Sbjct 215 CIFCPDVKARYESVQSCQQHMRDKQHCKLRRDPESMIELDDYYDYSPMY 263
> cel:K02A4.1 bcat-1; Branched Chain AminoTransferase family member
(bcat-1); K00826 branched-chain amino acid aminotransferase
[EC:2.6.1.42]
Length=415
Score = 30.8 bits (68), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 12/37 (32%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 145 RGWRHARVTDEGNLQLPSGQEVLHRSHAIFCRQRVRR 181
RGW H ++ G L++ G +VLH + +F + R
Sbjct 89 RGWHHPKIEPIGELKIHPGAKVLHYASELFEGMKAYR 125
> dre:563236 MGC173506, wu:fc26h11, wu:fk85d05; zgc:173506; K11367
chromodomain-helicase-DNA-binding protein 1 [EC:3.6.4.12]
Length=1693
Score = 30.8 bits (68), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 25/45 (55%), Gaps = 1/45 (2%)
Query 38 HFQLKW-DEEQQDLLHRFYDYKKSYYEILGRLPEVNNAQLVLPDN 81
HF ++W E+ LL Y+Y +E++ P++N +LPD+
Sbjct 1235 HFDIEWGKEDDSSLLIGIYEYGYGSWEMIKMDPDLNLTHKLLPDD 1279
> cel:T03F6.2 dnj-17; DNaJ domain (prokaryotic heat shock protein)
family member (dnj-17); K09506 DnaJ homolog subfamily A
member 5
Length=510
Score = 30.8 bits (68), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 10/32 (31%), Positives = 18/32 (56%), Gaps = 0/32 (0%)
Query 8 QEQQKQPRCLFCCRGFKGIRAALQHMQQQRHF 39
+E ++ P C+ C + FK + A L H ++H
Sbjct 310 EEGEELPYCVVCSKSFKTVNAKLNHENSKQHI 341
> cel:F14F4.3 mrp-5; Multidrug Resistance Protein family member
(mrp-5); K05668 ATP-binding cassette, subfamily C (CFTR/MRP),
member 5
Length=1400
Score = 30.4 bits (67), Expect = 4.2, Method: Composition-based stats.
Identities = 20/59 (33%), Positives = 28/59 (47%), Gaps = 1/59 (1%)
Query 89 ATSPLAAAKRTGRPLEAEAEVVAGED-EGDWEDCSSDEEGSANAASEQRRLEEMLQARG 146
ATSP PL+AE + ED +GD + SDEE N+ R ++ + A G
Sbjct 690 ATSPCGDGPAQPAPLDAEILRNSSEDLKGDADKLISDEEDMGNSTIAWRIYKQYIHAAG 748
> mmu:12648 Chd1, 4930525N21Rik, AI851787, AW555109, MGC141554;
chromodomain helicase DNA binding protein 1 (EC:3.6.4.12);
K11367 chromodomain-helicase-DNA-binding protein 1 [EC:3.6.4.12]
Length=1711
Score = 30.0 bits (66), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 13/45 (28%), Positives = 25/45 (55%), Gaps = 1/45 (2%)
Query 38 HFQLKW-DEEQQDLLHRFYDYKKSYYEILGRLPEVNNAQLVLPDN 81
HF + W E+ +LL Y+Y +E++ P+++ +LPD+
Sbjct 1246 HFDIDWGKEDDSNLLIGIYEYGYGSWEMIKMDPDLSLTHKILPDD 1290
> hsa:1105 CHD1, DKFZp686E2337; chromodomain helicase DNA binding
protein 1 (EC:3.6.4.12); K11367 chromodomain-helicase-DNA-binding
protein 1 [EC:3.6.4.12]
Length=1710
Score = 29.6 bits (65), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 13/45 (28%), Positives = 25/45 (55%), Gaps = 1/45 (2%)
Query 38 HFQLKW-DEEQQDLLHRFYDYKKSYYEILGRLPEVNNAQLVLPDN 81
HF + W E+ +LL Y+Y +E++ P+++ +LPD+
Sbjct 1248 HFDIDWGKEDDSNLLIGIYEYGYGSWEMIKMDPDLSLTHKILPDD 1292
> hsa:1015 CDH17, CDH16, FLJ26931, HPT-1, HPT1, MGC138218, MGC142024;
cadherin 17, LI cadherin (liver-intestine); K06811 cadherin
17, LI cadherin
Length=832
Score = 29.3 bits (64), Expect = 9.6, Method: Composition-based stats.
Identities = 17/66 (25%), Positives = 32/66 (48%), Gaps = 3/66 (4%)
Query 61 YYEILGRLPEVNNAQLVLPDNLSSSI---QDATSPLAAAKRTGRPLEAEAEVVAGEDEGD 117
YY+I+ +LP +NN +N + +I ++ + L AK L + + G+ E
Sbjct 164 YYQIVIQLPMINNVMYFQINNKTGAISLTREGSQELNPAKNPSYNLVISVKDMGGQSENS 223
Query 118 WEDCSS 123
+ D +S
Sbjct 224 FSDTTS 229
> hsa:170370 FAM170B, C10orf73; family with sequence similarity
170, member B
Length=283
Score = 29.3 bits (64), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 21/44 (47%), Gaps = 10/44 (22%)
Query 1 DLLRCLWQEQQKQP----------RCLFCCRGFKGIRAALQHMQ 34
DLL C QE ++ P RC+ CCR + A L+H Q
Sbjct 181 DLLECCLQELREPPDWLVTTNYGVRCVACCRVLPSLDALLEHAQ 224
Lambda K H
0.319 0.132 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5623228644
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40