bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8272_orf1
Length=61
Score E
Sequences producing significant alignments: (Bits) Value
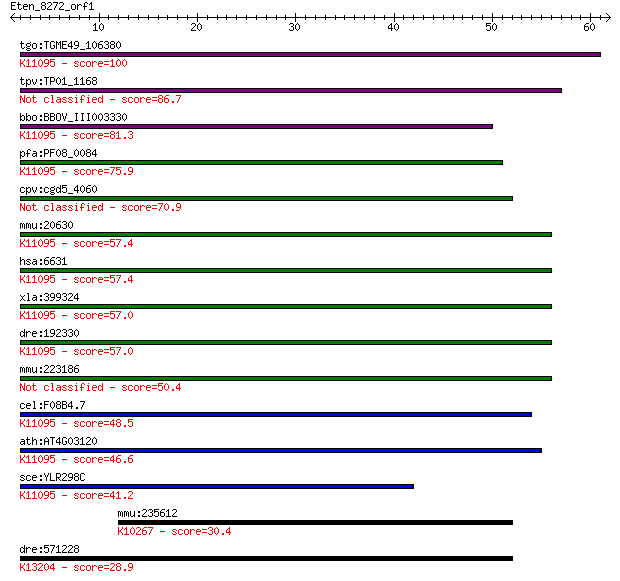
tgo:TGME49_106380 U1 small nuclear ribonucleoprotein, putative... 100 8e-22
tpv:TP01_1168 hypothetical protein 86.7 2e-17
bbo:BBOV_III003330 17.m07317; U1 zinc finger family protein; K... 81.3 7e-16
pfa:PF08_0084 RNA-binding protein (U1 snRNP-like), putative; K... 75.9 3e-14
cpv:cgd5_4060 hypothetical protein 70.9 9e-13
mmu:20630 Snrpc, Snrp1c, U1-C, U1C; U1 small nuclear ribonucle... 57.4 1e-08
hsa:6631 SNRPC, FLJ20302, U1C, Yhc1; small nuclear ribonucleop... 57.4 1e-08
xla:399324 snrpc, U1C, snrp1c; small nuclear ribonucleoprotein... 57.0 1e-08
dre:192330 snrpc, MGC109792, chunp6909, zgc:109792; small nucl... 57.0 2e-08
mmu:223186 Gm4822, EG223186; U1 small nuclear ribonucleoprotei... 50.4 1e-06
cel:F08B4.7 hypothetical protein; K11095 U1 small nuclear ribo... 48.5 5e-06
ath:AT4G03120 proline-rich family protein; K11095 U1 small nuc... 46.6 2e-05
sce:YLR298C YHC1; U1C; U1-C; K11095 U1 small nuclear ribonucle... 41.2 8e-04
mmu:235612 Fbxw19, E330023O18; F-box and WD-40 domain protein ... 30.4 1.4
dre:571228 znf326, MGC174183, si:dkey-238n5.2; zinc finger pro... 28.9 4.4
> tgo:TGME49_106380 U1 small nuclear ribonucleoprotein, putative
; K11095 U1 small nuclear ribonucleoprotein C
Length=215
Score = 100 bits (250), Expect = 8e-22, Method: Compositional matrix adjust.
Identities = 43/59 (72%), Positives = 49/59 (83%), Gaps = 0/59 (0%)
Query 2 YCEYCDIYLTHSSPAGRRQHASGRRHINQKVEYFQNLLRESTFQAPAFIDQAVAMRAIQ 60
YCEYCDIYLTHSSPAGRRQHA+GR+HINQK+EYFQNL+RE F P +D V RA+Q
Sbjct 5 YCEYCDIYLTHSSPAGRRQHATGRKHINQKIEYFQNLIREPDFMPPQQLDSTVVQRAVQ 63
> tpv:TP01_1168 hypothetical protein
Length=141
Score = 86.7 bits (213), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 36/55 (65%), Positives = 43/55 (78%), Gaps = 0/55 (0%)
Query 2 YCEYCDIYLTHSSPAGRRQHASGRRHINQKVEYFQNLLRESTFQAPAFIDQAVAM 56
YCEYC IYLTHSSPAGR+QH+ GR+HI+ KVEYFQ L+RE FQ P F+ Q +
Sbjct 5 YCEYCSIYLTHSSPAGRKQHSQGRKHISAKVEYFQRLVREQFFQPPVFLGQTPPI 59
> bbo:BBOV_III003330 17.m07317; U1 zinc finger family protein;
K11095 U1 small nuclear ribonucleoprotein C
Length=128
Score = 81.3 bits (199), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 33/48 (68%), Positives = 40/48 (83%), Gaps = 0/48 (0%)
Query 2 YCEYCDIYLTHSSPAGRRQHASGRRHINQKVEYFQNLLRESTFQAPAF 49
YCEYC IYLTHSSPAGR+QH+ GR+HI+ KVEY+Q L+RE FQ P +
Sbjct 5 YCEYCGIYLTHSSPAGRKQHSMGRKHISMKVEYYQRLVREHFFQPPPY 52
> pfa:PF08_0084 RNA-binding protein (U1 snRNP-like), putative;
K11095 U1 small nuclear ribonucleoprotein C
Length=235
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 34/49 (69%), Positives = 38/49 (77%), Gaps = 0/49 (0%)
Query 2 YCEYCDIYLTHSSPAGRRQHASGRRHINQKVEYFQNLLRESTFQAPAFI 50
YCEYCDIYLTHSSP GRRQH GR+HI+ K+EYFQNLLRE F+
Sbjct 5 YCEYCDIYLTHSSPVGRRQHIHGRKHISAKIEYFQNLLREEGITPQNFL 53
> cpv:cgd5_4060 hypothetical protein
Length=198
Score = 70.9 bits (172), Expect = 9e-13, Method: Composition-based stats.
Identities = 27/50 (54%), Positives = 39/50 (78%), Gaps = 0/50 (0%)
Query 2 YCEYCDIYLTHSSPAGRRQHASGRRHINQKVEYFQNLLRESTFQAPAFID 51
YC+YCDIYLTHSS GR+QH GR+HIN K+++++N+++ +F AP D
Sbjct 5 YCDYCDIYLTHSSTNGRKQHNLGRKHINNKIDHYKNVIKSPSFSAPLMFD 54
> mmu:20630 Snrpc, Snrp1c, U1-C, U1C; U1 small nuclear ribonucleoprotein
C; K11095 U1 small nuclear ribonucleoprotein C
Length=159
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 25/54 (46%), Positives = 34/54 (62%), Gaps = 3/54 (5%)
Query 2 YCEYCDIYLTHSSPAGRRQHASGRRHINQKVEYFQNLLRESTFQAPAFIDQAVA 55
YC+YCD YLTH SP+ R+ H SGR+H +Y+Q + E QA + ID+ A
Sbjct 5 YCDYCDTYLTHDSPSVRKTHCSGRKHKENVKDYYQKWMEE---QAQSLIDKTTA 55
> hsa:6631 SNRPC, FLJ20302, U1C, Yhc1; small nuclear ribonucleoprotein
polypeptide C; K11095 U1 small nuclear ribonucleoprotein
C
Length=159
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 25/54 (46%), Positives = 34/54 (62%), Gaps = 3/54 (5%)
Query 2 YCEYCDIYLTHSSPAGRRQHASGRRHINQKVEYFQNLLRESTFQAPAFIDQAVA 55
YC+YCD YLTH SP+ R+ H SGR+H +Y+Q + E QA + ID+ A
Sbjct 5 YCDYCDTYLTHDSPSVRKTHCSGRKHKENVKDYYQKWMEE---QAQSLIDKTTA 55
> xla:399324 snrpc, U1C, snrp1c; small nuclear ribonucleoprotein
polypeptide C; K11095 U1 small nuclear ribonucleoprotein
C
Length=159
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 25/54 (46%), Positives = 34/54 (62%), Gaps = 3/54 (5%)
Query 2 YCEYCDIYLTHSSPAGRRQHASGRRHINQKVEYFQNLLRESTFQAPAFIDQAVA 55
YC+YCD YLTH SP+ R+ H SGR+H +Y+Q + E QA + ID+ A
Sbjct 5 YCDYCDTYLTHDSPSVRKTHCSGRKHKENVKDYYQKWMEE---QAQSLIDKTTA 55
> dre:192330 snrpc, MGC109792, chunp6909, zgc:109792; small nuclear
ribonucleoprotein polypeptide C; K11095 U1 small nuclear
ribonucleoprotein C
Length=159
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 25/54 (46%), Positives = 34/54 (62%), Gaps = 3/54 (5%)
Query 2 YCEYCDIYLTHSSPAGRRQHASGRRHINQKVEYFQNLLRESTFQAPAFIDQAVA 55
YC+YCD YLTH SP+ R+ H SGR+H +Y+Q + E QA + ID+ A
Sbjct 5 YCDYCDTYLTHDSPSVRKTHCSGRKHKENVKDYYQKWMEE---QAQSLIDKTTA 55
> mmu:223186 Gm4822, EG223186; U1 small nuclear ribonucleoprotein
1C pseudogene
Length=159
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 24/54 (44%), Positives = 31/54 (57%), Gaps = 3/54 (5%)
Query 2 YCEYCDIYLTHSSPAGRRQHASGRRHINQKVEYFQNLLRESTFQAPAFIDQAVA 55
YC YCD+YLTH+SP+ R+ H SGR+H E Q + E Q ID+ A
Sbjct 5 YCYYCDMYLTHNSPSMRKTHCSGRKHKENVKENCQKWMEE---QVQTLIDKPTA 55
> cel:F08B4.7 hypothetical protein; K11095 U1 small nuclear ribonucleoprotein
C
Length=142
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 31/52 (59%), Gaps = 3/52 (5%)
Query 2 YCEYCDIYLTHSSPAGRRQHASGRRHINQKVEYFQNLLRESTFQAPAFIDQA 53
YC+YCD +LTH SP+ R+ H GR+H + ++Q + + QA +DQ
Sbjct 5 YCDYCDTFLTHDSPSVRKTHNGGRKHKDNVRMFYQKWMED---QAQKLVDQT 53
> ath:AT4G03120 proline-rich family protein; K11095 U1 small nuclear
ribonucleoprotein C
Length=207
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 22/53 (41%), Positives = 31/53 (58%), Gaps = 3/53 (5%)
Query 2 YCEYCDIYLTHSSPAGRRQHASGRRHINQKVEYFQNLLRESTFQAPAFIDQAV 54
YC+YCD YLTH SP+ R+QH +G +H Y+Q + T + IDQ +
Sbjct 5 YCDYCDTYLTHDSPSVRKQHNAGYKHKANVRIYYQQFEEQQTQ---SLIDQRI 54
> sce:YLR298C YHC1; U1C; U1-C; K11095 U1 small nuclear ribonucleoprotein
C
Length=231
Score = 41.2 bits (95), Expect = 8e-04, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 25/40 (62%), Gaps = 0/40 (0%)
Query 2 YCEYCDIYLTHSSPAGRRQHASGRRHINQKVEYFQNLLRE 41
YCEYC YLTH + + R+ H G+ H+ +Y++N R+
Sbjct 5 YCEYCHSYLTHDTLSVRKSHLVGKNHLRITADYYRNKARD 44
> mmu:235612 Fbxw19, E330023O18; F-box and WD-40 domain protein
19; K10267 F-box and WD-40 domain protein 12/13/14/15/16/17/19
Length=466
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 21/41 (51%), Gaps = 1/41 (2%)
Query 12 HSSPAGRRQHASGR-RHINQKVEYFQNLLRESTFQAPAFID 51
H SP + G+ RH+ KV Y +LLR S F AP D
Sbjct 240 HCSPQKKWVLLIGKQRHVLTKVFYMSSLLRTSEFSAPVSTD 280
> dre:571228 znf326, MGC174183, si:dkey-238n5.2; zinc finger protein
326; K13204 zinc finger protein 326
Length=451
Score = 28.9 bits (63), Expect = 4.4, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 25/50 (50%), Gaps = 1/50 (2%)
Query 2 YCEYCDIYLTHSSPAGRRQHASGRRHINQKVEYFQNLLRESTFQAPAFID 51
YC CD+++ + +QH H+ KV Y + L R+S A A I+
Sbjct 355 YCVACDVHIP-VLFSSVQQHLHSLTHLKSKVAYKEQLKRDSVVTAKAIIN 403
Lambda K H
0.326 0.135 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2064920148
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40