bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_8309_orf1
Length=105
Score E
Sequences producing significant alignments: (Bits) Value
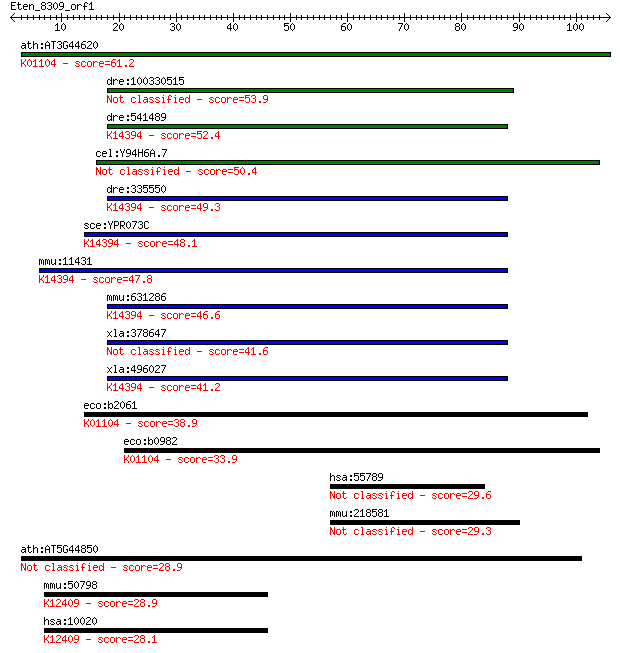
ath:AT3G44620 protein tyrosine phosphatase; K01104 protein-tyr... 61.2 7e-10
dre:100330515 acid phosphatase 1, soluble-like 53.9 1e-07
dre:541489 acp1, im:6910498, zgc:110844; acid phosphatase 1, s... 52.4 3e-07
cel:Y94H6A.7 hypothetical protein 50.4 1e-06
dre:335550 fj21b03; wu:fj21b03; K14394 low molecular weight ph... 49.3 3e-06
sce:YPR073C LTP1; Ltp1p (EC:3.1.3.48 3.1.3.2); K14394 low mole... 48.1 7e-06
mmu:11431 Acp1, 4632432E04Rik, AI427468, Acp-1, LMW-PTP, MGC10... 47.8 8e-06
mmu:631286 low molecular weight phosphotyrosine protein phosph... 46.6 2e-05
xla:378647 acp1-a, LMW-PTP, MGC80022, XLPTP1, acp1, acp1a, haa... 41.6 5e-04
xla:496027 acp1-b, acp1b, haap, lmw-ptp, ptp1a, xlptp1; acid p... 41.2 8e-04
eco:b2061 wzb, ECK2055, JW2046; protein-tyrosine phosphatase (... 38.9 0.004
eco:b0982 etp, ECK0973, JW5132, yccY; phosphotyrosine-protein ... 33.9 0.11
hsa:55789 DEPDC1B, BRCC3, FLJ11252, XTP1; DEP domain containin... 29.6 2.6
mmu:218581 Depdc1b, 9830132O03, AW260467, XTP1; DEP domain con... 29.3 2.8
ath:AT5G44850 hypothetical protein 28.9 4.2
mmu:50798 Gne, 2310066H07Rik, DMRV, GLCNE, IBM2, NM, Uae1; glu... 28.9 4.3
hsa:10020 GNE, DMRV, GLCNE, IBM2, NM, Uae1; glucosamine (UDP-N... 28.1 6.8
> ath:AT3G44620 protein tyrosine phosphatase; K01104 protein-tyrosine
phosphatase [EC:3.1.3.48]
Length=239
Score = 61.2 bits (147), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 40/114 (35%), Positives = 61/114 (53%), Gaps = 13/114 (11%)
Query 3 HTEGDPAMGEMIRAARKRGVDLSPHRARQVRKTDFEEFDLVVAMDKSNLRNLAAL----- 57
+ EG+ A M AA++RG++++ +R ++ +DF EFDL++AMD N ++
Sbjct 120 YHEGNMADPRMRSAAKRRGIEIT-SLSRPIKASDFREFDLILAMDDQNKEDILKAYNVWK 178
Query 58 ----CPPDKRHKLKLLIRDYAPECGTEEVPDPYYEG--GHLNVFDLIEKGVEGL 105
PPD K+KL+ Y + + VPDPYY G G V DL+E E L
Sbjct 179 ARGNFPPDADKKVKLMC-SYCKKHNDKFVPDPYYGGAQGFEKVLDLLEDACESL 231
> dre:100330515 acid phosphatase 1, soluble-like
Length=139
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 32/72 (44%), Positives = 43/72 (59%), Gaps = 3/72 (4%)
Query 18 RKRGVDLSPHRARQVRKTDFEEFDLVVAMDKSNLRNLAALCPPDKRHKLKL-LIRDYAPE 76
RK G++ + HRARQV K DF FD ++ MD+SNLR+L K K K+ L+ Y PE
Sbjct 65 RKHGIE-TDHRARQVTKDDFMSFDYILCMDESNLRDLNKKASSVKNSKAKIELLGSYDPE 123
Query 77 CGTEEVPDPYYE 88
+ DPYY+
Sbjct 124 -KQLIIQDPYYD 134
> dre:541489 acp1, im:6910498, zgc:110844; acid phosphatase 1,
soluble; K14394 low molecular weight phosphotyrosine protein
phosphatase [EC:3.1.3.2 3.1.3.48]
Length=158
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 31/71 (43%), Positives = 42/71 (59%), Gaps = 3/71 (4%)
Query 18 RKRGVDLSPHRARQVRKTDFEEFDLVVAMDKSNLRNLAALCPPDKRHKLKL-LIRDYAPE 76
RK G++ + HRARQV K DF FD ++ MD+SNLR+L + K K+ L+ Y PE
Sbjct 65 RKHGIE-TDHRARQVTKDDFMSFDYILCMDESNLRDLNKKASSVENSKAKIELLGSYDPE 123
Query 77 CGTEEVPDPYY 87
+ DPYY
Sbjct 124 -KQLIIQDPYY 133
> cel:Y94H6A.7 hypothetical protein
Length=104
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/91 (31%), Positives = 45/91 (49%), Gaps = 3/91 (3%)
Query 16 AARKRGVDLSPHRARQVRKTDFEEFDLVVAMDKSNLRNLAALC---PPDKRHKLKLLIRD 72
A +K G+ HRAR DF +FD + MD N+ +L + P +R L++
Sbjct 3 ALKKYGIKDYQHRARVTSPDDFRKFDYIFGMDDQNIEDLQEIARKVPKTERKAEILMLGV 62
Query 73 YAPECGTEEVPDPYYEGGHLNVFDLIEKGVE 103
G EVPDPYYE G +++++ V+
Sbjct 63 QDVMAGKREVPDPYYESGSKQFDEVLQQCVK 93
> dre:335550 fj21b03; wu:fj21b03; K14394 low molecular weight
phosphotyrosine protein phosphatase [EC:3.1.3.2 3.1.3.48]
Length=158
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 31/71 (43%), Positives = 40/71 (56%), Gaps = 3/71 (4%)
Query 18 RKRGVDLSPHRARQVRKTDFEEFDLVVAMDKSNLRNLAALCPPDKRHKLKL-LIRDYAPE 76
+K GV + H ARQV K DF FD ++ MD+SNLR+L K K K+ L+ Y PE
Sbjct 65 KKHGVSMR-HVARQVTKDDFMSFDYILCMDESNLRDLNKKASSVKNSKAKIELLGSYDPE 123
Query 77 CGTEEVPDPYY 87
+ DPYY
Sbjct 124 -KKLIIQDPYY 133
> sce:YPR073C LTP1; Ltp1p (EC:3.1.3.48 3.1.3.2); K14394 low molecular
weight phosphotyrosine protein phosphatase [EC:3.1.3.2
3.1.3.48]
Length=161
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 22/76 (28%), Positives = 44/76 (57%), Gaps = 4/76 (5%)
Query 14 IRAARKRGVDLSPHRARQVRKTDFEEFDLVVAMDKSNLRNLAALCPPDKRHKLKLLIRDY 73
+ ++ GV ++ H+ +Q++ F+E+D ++ MD+SN+ NL + P + K+ L D+
Sbjct 63 VSICKQHGVKIN-HKGKQIKTKHFDEYDYIIGMDESNINNLKKIQPEGSKAKV-CLFGDW 120
Query 74 APECGTEE--VPDPYY 87
GT + + DP+Y
Sbjct 121 NTNDGTVQTIIEDPWY 136
> mmu:11431 Acp1, 4632432E04Rik, AI427468, Acp-1, LMW-PTP, MGC103115,
MGC132904; acid phosphatase 1, soluble (EC:3.1.3.2 3.1.3.48);
K14394 low molecular weight phosphotyrosine protein
phosphatase [EC:3.1.3.2 3.1.3.48]
Length=158
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 30/83 (36%), Positives = 43/83 (51%), Gaps = 3/83 (3%)
Query 6 GDPAMGEMIRAARKRGVDLSPHRARQVRKTDFEEFDLVVAMDKSNLRNLAALCPPDKRHK 65
G P + R G+ + H+ARQ+ K DF FD ++ MD+SNLR+L K K
Sbjct 53 GRPPDPRAVSCLRNHGIS-TAHKARQITKEDFATFDYILCMDESNLRDLNRKSNQVKNCK 111
Query 66 LKL-LIRDYAPECGTEEVPDPYY 87
K+ L+ Y P+ + DPYY
Sbjct 112 AKIELLGSYDPQ-KQLIIEDPYY 133
> mmu:631286 low molecular weight phosphotyrosine protein phosphatase-like;
K14394 low molecular weight phosphotyrosine protein
phosphatase [EC:3.1.3.2 3.1.3.48]
Length=158
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 29/71 (40%), Positives = 40/71 (56%), Gaps = 3/71 (4%)
Query 18 RKRGVDLSPHRARQVRKTDFEEFDLVVAMDKSNLRNLAALCPPDKRHKLKL-LIRDYAPE 76
RK G+ + H ARQ+ K DF FD ++ MD+SNLR+L K K K+ L+ Y P+
Sbjct 65 RKHGIHMQ-HIARQITKEDFATFDYILCMDESNLRDLNRKSNQVKNCKAKIELLGSYDPQ 123
Query 77 CGTEEVPDPYY 87
+ DPYY
Sbjct 124 -KQLIIEDPYY 133
> xla:378647 acp1-a, LMW-PTP, MGC80022, XLPTP1, acp1, acp1a, haap,
ptp1a, ptp1a-A; acid phosphatase 1, soluble
Length=159
Score = 41.6 bits (96), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 41/71 (57%), Gaps = 3/71 (4%)
Query 18 RKRGVDLSPHRARQVRKTDFEEFDLVVAMDKSNLRNLAALCPPDKRHKLKL-LIRDYAPE 76
+K G+ +S H ARQ+ + DF +D ++ MD+SNL++L + K K+ L+ Y P+
Sbjct 66 KKHGIPMS-HTARQITRDDFLSYDYILCMDESNLQDLKRRGSQVQNCKAKIELLGSYDPQ 124
Query 77 CGTEEVPDPYY 87
+ DPYY
Sbjct 125 -KQLIIQDPYY 134
> xla:496027 acp1-b, acp1b, haap, lmw-ptp, ptp1a, xlptp1; acid
phosphatase 1, soluble; K14394 low molecular weight phosphotyrosine
protein phosphatase [EC:3.1.3.2 3.1.3.48]
Length=158
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 39/71 (54%), Gaps = 3/71 (4%)
Query 18 RKRGVDLSPHRARQVRKTDFEEFDLVVAMDKSNLRNLAALCPPDKRHKLKL-LIRDYAPE 76
+ G+ +S H ARQ+ DF +D ++ MD+SNLR+L + K K+ L+ Y P+
Sbjct 65 KNHGIPMS-HTARQITSNDFLSYDYILCMDESNLRDLKKKGGQVQNCKAKIELLGSYDPQ 123
Query 77 CGTEEVPDPYY 87
+ DPYY
Sbjct 124 -KKLIIEDPYY 133
> eco:b2061 wzb, ECK2055, JW2046; protein-tyrosine phosphatase
(EC:3.1.3.48); K01104 protein-tyrosine phosphatase [EC:3.1.3.48]
Length=147
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 26/90 (28%), Positives = 46/90 (51%), Gaps = 7/90 (7%)
Query 14 IRAARKRGVDLSPHRARQVRKTDFEEFDLVVAMDKSNLRNLAALCPPDKRHKLKLLIRDY 73
I A + + L H ARQ+ + +DL++ M+K ++ L + P+ R K+ +L +
Sbjct 50 ISVAAEHQLSLEGHCARQISRRLCRNYDLILTMEKRHIERLCEM-APEMRGKV-MLFGHW 107
Query 74 APECGTEEVPDPYYEG--GHLNVFDLIEKG 101
EC E+PDPY + V+ L+E+
Sbjct 108 DNEC---EIPDPYRKSRETFAAVYTLLERS 134
> eco:b0982 etp, ECK0973, JW5132, yccY; phosphotyrosine-protein
phosphatase; K01104 protein-tyrosine phosphatase [EC:3.1.3.48]
Length=148
Score = 33.9 bits (76), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 21/85 (24%), Positives = 42/85 (49%), Gaps = 7/85 (8%)
Query 21 GVDLSPHRARQVRKTDFEEFDLVVAMDKSNLRNLAALCPPDKRHKLKLLIRDYAPECGTE 80
GV L H R++ +DL++AM+ ++ + A+ P+ R K L + +
Sbjct 61 GVSLEGHAGRKLTAEMARNYDLILAMESEHIAQVTAIA-PEVRGKTML----FGQWLEQK 115
Query 81 EVPDPYYEG--GHLNVFDLIEKGVE 103
E+PDPY + +V+ ++E+ +
Sbjct 116 EIPDPYRKSQDAFEHVYGMLERASQ 140
> hsa:55789 DEPDC1B, BRCC3, FLJ11252, XTP1; DEP domain containing
1B
Length=467
Score = 29.6 bits (65), Expect = 2.6, Method: Composition-based stats.
Identities = 13/27 (48%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 57 LCPPDKRHKLKLLIRDYAPECGTEEVP 83
L PP+ R KL+LL+R A C +E+P
Sbjct 318 LLPPENRRKLQLLMRMMARICLNKEMP 344
> mmu:218581 Depdc1b, 9830132O03, AW260467, XTP1; DEP domain containing
1B
Length=529
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 21/33 (63%), Gaps = 1/33 (3%)
Query 57 LCPPDKRHKLKLLIRDYAPECGTEEVPDPYYEG 89
L PP+ R KL+LL+R A C +E+P P +G
Sbjct 318 LLPPENRRKLQLLMRMMARICLNKEMP-PLSDG 349
> ath:AT5G44850 hypothetical protein
Length=331
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 28/108 (25%), Positives = 46/108 (42%), Gaps = 12/108 (11%)
Query 3 HTEGDPAMGEMIRAARKRGVDLSPHRARQVRKTDFEEF-DLVVAMDKSN-LRNLAALCPP 60
HT D + + + +DL+ + +F +L++ D+S+ L L L
Sbjct 167 HTYADDKLPRSLSSVLFLELDLADEMLLRCSAINFSRLTELIICPDESDWLEPLMLLL-- 224
Query 61 DKRHKLKLLIRDYAPECGTEEVPDPYYE--------GGHLNVFDLIEK 100
K KLK L DY P E++P + E HL +F+ +EK
Sbjct 225 GKSPKLKKLSVDYGPTENPEDLPLSWNEPSSVPGCLSSHLEIFEWVEK 272
> mmu:50798 Gne, 2310066H07Rik, DMRV, GLCNE, IBM2, NM, Uae1; glucosamine
(EC:5.1.3.14 2.7.1.60); K12409 bifunctional UDP-N-acetylglucosamine
2-epimerase / N-acetylmannosamine kinase
[EC:5.1.3.14 2.7.1.60]
Length=722
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query 7 DPAMGEMIRAARKRGVDLSPHRARQVRKTDFEEFDLVVA 45
D EM+R RK+G++ P+ R V+ F++F +VA
Sbjct 255 DAGSKEMVRVMRKKGIEHHPN-FRAVKHVPFDQFIQLVA 292
> hsa:10020 GNE, DMRV, GLCNE, IBM2, NM, Uae1; glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine
kinase (EC:5.1.3.14
2.7.1.60); K12409 bifunctional UDP-N-acetylglucosamine 2-epimerase
/ N-acetylmannosamine kinase [EC:5.1.3.14 2.7.1.60]
Length=753
Score = 28.1 bits (61), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query 7 DPAMGEMIRAARKRGVDLSPHRARQVRKTDFEEFDLVVA 45
D EM+R RK+G++ P+ R V+ F++F +VA
Sbjct 286 DAGSKEMVRVMRKKGIEHHPN-FRAVKHVPFDQFIQLVA 323
Lambda K H
0.318 0.140 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2017521068
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40